6VCZ
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) | | Descriptor: | 2-METHOXYETHANOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
5TZB
 
 | | Burkholderia sp. beta-aminopeptidase | | Descriptor: | CALCIUM ION, D-aminopeptidase | | Authors: | McGowan, S, Drinkwater, N, John, M, Dumsday, G. | | Deposit date: | 2016-11-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Crystal structure of a beta-aminopeptidase from an Australian Burkholderia sp.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1GPK
 
 | | Structure of Acetylcholinesterase Complex with (+)-Huperzine A at 2.1A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Dvir, H, Harel, M, Chetrit, M, Silman, I, Sussman, J.L. | | Deposit date: | 2001-11-05 | | Release date: | 2002-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Torpedo Californica Acetylcholinesterase Complexed with (+)-Huperzine a and (-)-Huperzine B: Structural Evidence for an Active Site Rearrangement
Biochemistry, 41, 2002
|
|
4O1D
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | Descriptor: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
1GPW
 
 | | Structural evidence for ammonia tunneling across the (beta/alpha)8 barrel of the imidazole glycerol phosphate synthase bienzyme complex. | | Descriptor: | AMIDOTRANSFERASE HISH, HISF PROTEIN, PHOSPHATE ION | | Authors: | Walker, M, Beismann-Driemeyer, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Evidence for Ammonia Tunneling Across the (Beta Alpha)(8) Barrel of the Imidazole Glycerol Phosphate Synthase Bienzyme Complex.
Structure, 10, 2002
|
|
6VAR
 
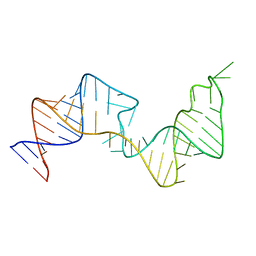 | | 61 nt human Hepatitis B virus epsilon pre-genomic RNA | | Descriptor: | RNA (61-MER) | | Authors: | LeBlanc, R.M, Kasprzak, W.K, Longhini, A.P, Abulwerdi, F, Ginocchio, S, Shields, B, Nyman, J, Svirydava, M, Del Vecchio, C, Ivanic, J, Schneekloth, J.S, Dayie, T.K, Shapiro, B.A, Le Grice, S.F.J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural insights of the conserved "priming loop" of hepatitis B virus pre-genomic RNA.
J.Biomol.Struct.Dyn., 2021
|
|
6VE2
 
 | | Tetradecameric PilQ bound by TsaP heptamer from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ, LysM domain-containing protein | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6V6B
 
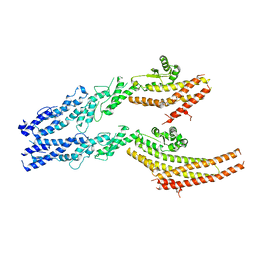 | | Structures of GCP2 and GCP3 in the native human gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 2, Gamma-tubulin complex component 3 | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
8DFW
 
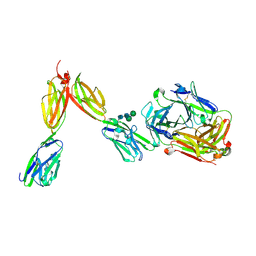 | | Crystal Structure of Human BTN2A1 in Complex With Vgamma9-Vdelta2 T Cell Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
2C0F
 
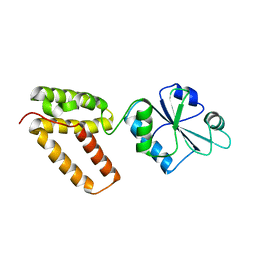 | | Structure of Wind Y53F mutant | | Descriptor: | WINDBEUTEL PROTEIN | | Authors: | Sevvana, M, Ma, Q, Barnewitz, K, Guo, C, Soling, H.-D, Ferrari, D.M, Sheldrick, G.M. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Elucidation of the Pdi-Related Chaperone Wind with the Help of Mutants.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1GYC
 
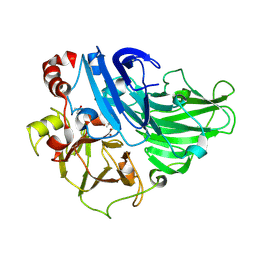 | | CRYSTAL STRUCTURE DETERMINATION AT ROOM TEMPERATURE OF A LACCASE FROM TRAMETES VERSICOLOR IN ITS OXIDISED FORM CONTAINING A FULL COMPLEMENT OF COPPER IONS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Choinowski, T, Antorini, M, Piontek, K. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Laccase from the Fungus Trametes Versicolor at 1.90-A Resolution Containing a Full Complement of Coppers.
J.Biol.Chem., 277, 2002
|
|
3CYN
 
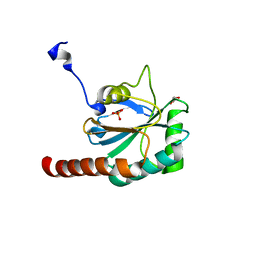 | | The structure of human GPX8 | | Descriptor: | GLYCEROL, Probable glutathione peroxidase 8, SULFATE ION | | Authors: | Kavanagh, K.L, Johansson, C, Yue, W.W, Kochan, G, Pike, A.C.W, Murray, J, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human GPX8
To be Published
|
|
1HH0
 
 | | Filamentous Bacteriophage PH75 | | Descriptor: | PH75 INOVIRUS MAJOR COAT PROTEIN | | Authors: | Pederson, D.M, Welsh, L.C, Marvin, D.A, Sampson, M, Perham, R.N, Yu, M, Slater, M.R. | | Deposit date: | 2000-12-17 | | Release date: | 2001-06-01 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (2.4 Å) | | Cite: | The Protein Capsid of Filamentous Bacteriophage Ph75 from Thermus Thermophilus
J.Mol.Biol., 309, 2001
|
|
2C5Y
 
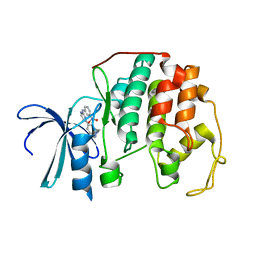 | | DIFFERENTIAL BINDING OF INHIBITORS TO ACTIVE AND INACTIVE CDK2 PROVIDES INSIGHTS FOR DRUG DESIGN | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2MDB
 
 | | Tachyplesin I in the presence of lipopolysaccharide | | Descriptor: | Tachyplesin-1 | | Authors: | Kushibiki, T, Kamiya, M, Aizawa, T, Kumaki, Y, Kikukawa, T, Mizuguchi, M, Demura, M, Kawabata, S.I, Kawano, K. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide.
Biochim.Biophys.Acta, 1844, 2014
|
|
471D
 
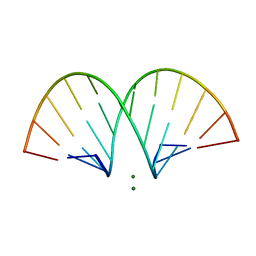 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
1H21
 
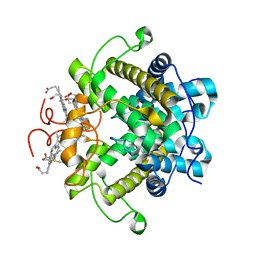 | | A novel iron centre in the split-Soret cytochrome c from Desulfovibrio desulfuricans ATCC 27774 | | Descriptor: | HEME C, SPLIT-SORET CYTOCHROME C | | Authors: | Abreu, I.A, Lourenco, A.I, Xavier, A.V, Legall, J, Coelho, A.V, Matias, P.M, Pinto, D.M, Carrondo, M.A, Teixeira, M, Saraiva, L.M. | | Deposit date: | 2002-07-30 | | Release date: | 2003-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Iron Centre in the Split-Soret Cytochrome C from Desulfovibrio Desulfuricans Atcc 27774
J.Biol.Inorg.Chem., 8, 2003
|
|
4NST
 
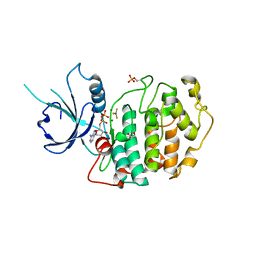 | | Crystal structure of human Cdk12/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Boesken, C.A, Farnung, L, Anand, K, Geyer, M. | | Deposit date: | 2013-11-29 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure and substrate specificity of human Cdk12/Cyclin K.
Nat Commun, 5, 2014
|
|
2MF6
 
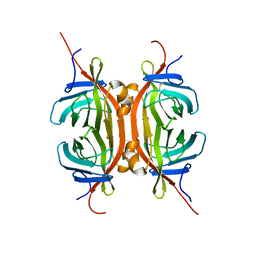 | | Solution NMR structure of Chimeric Avidin, ChiAVD(I117Y), in the biotin bound form | | Descriptor: | Avidin, Avidin-related protein 4/5 | | Authors: | Tossavainen, H, Kukkurainen, S, Maatta, J.A.E, Pihlajamaa, T, Hytonen, V.P, Kulomaa, M.S, Permi, P. | | Deposit date: | 2013-10-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Chimeric Avidin - NMR Structure and Dynamics of a 56 kDa Homotetrameric Thermostable Protein
Plos One, 9, 2014
|
|
4KE8
 
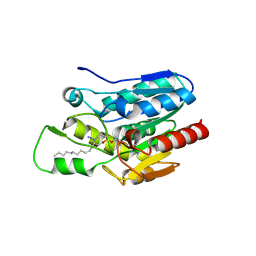 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with monopalmitoyl glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, tetradecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
6V7P
 
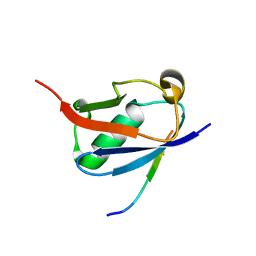 | | Crystal structure of SUMO1 in complex with PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
5RE6
 
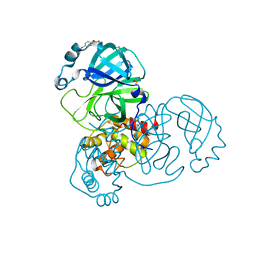 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z54571979 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-{4-[(pyrimidin-2-yl)oxy]phenyl}acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2LYC
 
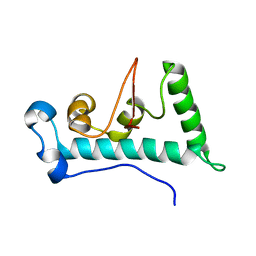 | | Structure of C-terminal domain of Ska1 | | Descriptor: | Spindle and kinetochore-associated protein 1 homolog | | Authors: | Boeszoermenyi, A, Schmidt, J.C, Markus, M, Oberer, M, Cheeseman, I.M, Wagner, G, Arthanari, H. | | Deposit date: | 2012-09-14 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments.
Dev.Cell, 23, 2012
|
|
5REM
 
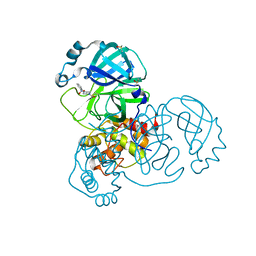 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0103016 | | Descriptor: | 1 1-(4-(2-nitrophenyl)piperazin-1-yl)ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RF3
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1741970824 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, pyrimidin-5-amine | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
