5VEG
 
 | | Structure of a Short-Chain Flavodoxin Associated with a Non-Canonical PDU Bacterial Microcompartment | | Descriptor: | CADMIUM ION, FLAVIN MONONUCLEOTIDE, Flavodoxin, ... | | Authors: | Sutter, M, Plegaria, J.S, Kerfeld, C.A. | | Deposit date: | 2017-04-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Functional Characterization of a Short-Chain Flavodoxin Associated with a Noncanonical 1,2-Propanediol Utilization Bacterial Microcompartment.
Biochemistry, 56, 2017
|
|
3NTM
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium crystallized in the absence of zinc, partial occupancy of CuB | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Sendovski, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2010-07-05 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First structures of an active bacterial tyrosinase reveal copper plasticity
J.Mol.Biol., 405, 2011
|
|
6KMF
 
 | | FimA type V pilus from P.gingivalis | | Descriptor: | Major fimbrium subunit FimA type-1 | | Authors: | Shibata, S, Shoji, M, Matsunami, H, Matthews, M, Imada, K, Nakayama, K, Wolf, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
1MKV
 
 | |
1D87
 
 | |
5D6H
 
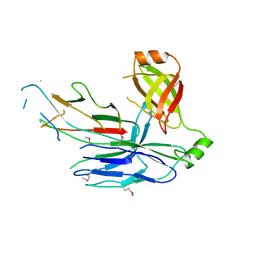 | | Crystal structure of CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Csu biofilm-mediating pili of Acinetobacter baumannii | | Descriptor: | CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-04 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis.
Plos Pathog., 11, 2015
|
|
1UVX
 
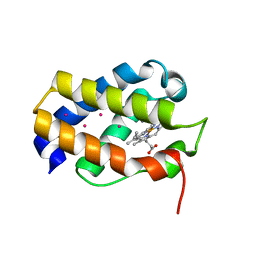 | | Heme-ligand tunneling on group I truncated hemoglobins | | Descriptor: | CYANIDE ION, GLOBIN LI637, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Milani, M, Pesce, A, Ouellet, Y, Dewilde, S, Friedman, J, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2004-01-27 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heme-Ligand Tunneling in Group I Truncated Hemoglobins
J.Biol.Chem., 279, 2004
|
|
5R0X
 
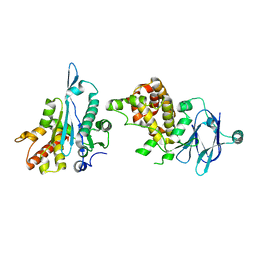 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 11, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1E
 
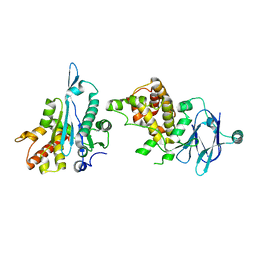 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 29, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1U
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry B03, DMSO-free | | Descriptor: | 2-[(1R)-1-aminopropyl]phenol, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R15
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 20, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R27
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry G03, DMSO-free | | Descriptor: | 3-ethoxybenzene-1-carboximidamide, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.029 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2M
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 10, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.008 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1V25
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, long-chain-fatty-acid-CoA synthetase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
5R1G
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 31, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R32
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 26, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1Z
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry D11, DMSO-free | | Descriptor: | 3-(1,3-thiazol-2-yl)propanoic acid, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.067 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3K
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 44, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3Z
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 59, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.067 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5VKF
 
 | | RHCC in complex with Naphthalene | | Descriptor: | NAPHTHALENE, SULFATE ION, Tetrabrachion | | Authors: | McDougall, M, Stetefeld, J. | | Deposit date: | 2017-04-21 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Proteinaceous Nano container Encapsulate Polycyclic Aromatic Hydrocarbons.
Sci Rep, 9, 2019
|
|
5R2E
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 01, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2U
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 18, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.138 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1V3U
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase in apo form | | Descriptor: | CHLORIDE ION, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
5R39
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 33, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3R
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 51, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.979 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
