8H51
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5P
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6N
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (T21I) in complex with protease inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H57
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) A193P Mutant in Complex with Inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
2Q2H
 
 | | Crystal structure of the protein secretion chaperone CsaA from Agrobacterium tumefaciens with a genetically fused phage-display derived peptide substrate at the N-terminus. | | Descriptor: | ACETATE ION, CITRIC ACID, Secretion chaperone, ... | | Authors: | Feldman, A.R, Shapova, Y.A, Paetzel, M. | | Deposit date: | 2007-05-28 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phage display and crystallographic analysis reveals potential substrate/binding site interactions in the protein secretion chaperone CsaA from Agrobacterium tumefaciens.
J.Mol.Biol., 379, 2008
|
|
8H5A
 
 | |
2Q3X
 
 | | The RIM1alpha C2B domain | | Descriptor: | CHLORIDE ION, Regulating synaptic membrane exocytosis protein 1, SODIUM ION, ... | | Authors: | Guan, R, Dai, H, Tomchick, D.R, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2007-05-30 | | Release date: | 2007-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the RIM1alpha C(2)B Domain at 1.7 A Resolution.
Biochemistry, 46, 2007
|
|
8H7W
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (S144A) in complex with protease inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H82
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
6TY5
 
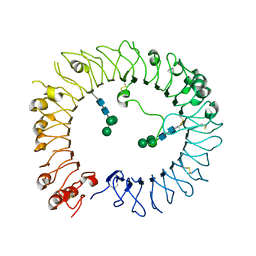 | | Crystal structure of human TLR8 in complex with Compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-7-(7-methyl-2-piperidin-4-yl-indazol-5-yl)furo[3,2-c]pyridin-4-one, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Target-Based Identification and Optimization of 5-Indazol-5-yl Pyridones as Toll-like Receptor 7 and 8 Antagonists Using a Biochemical TLR8 Antagonist Competition Assay.
J.Med.Chem., 63, 2020
|
|
6GKG
 
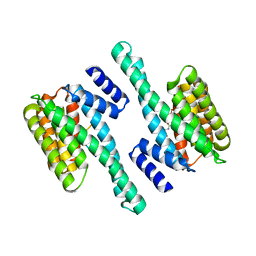 | |
8H58
 
 | | Crystal structure of YhaJ effector binding domain | | Descriptor: | HTH-type transcriptional regulator YhaJ, SODIUM ION | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structural basis of transcription factor YhaJ for DNT detection.
Iscience, 26, 2023
|
|
2CM7
 
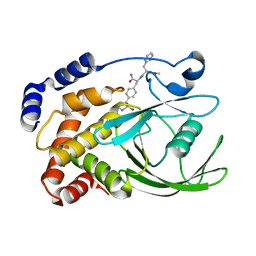 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | ISOTHIAZOLIDINONE ANALOG, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
8H7Q
 
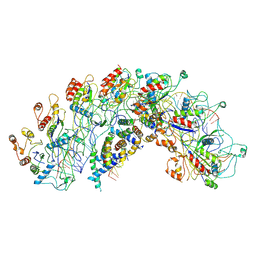 | | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution | | Descriptor: | CRISPR RNA, CRISPR associated protein Cas11b, CRISPR associated protein Cas5, ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution
To Be Published
|
|
6IMW
 
 | | The complex structure of endo-beta-1,2-glucanase mutant (E262Q) from Talaromyces funiculosus with beta-1,2-glucan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Endo-beta-1,2-glucanase, ... | | Authors: | Tanaka, N, Nakajima, M, Narukawa-Nara, M, Matsunaga, H, Kamisuki, S, Aramasa, H, Takahashi, Y, Sugimoto, N, Abe, K, Miyanaga, A, Yamashita, T, Sugawara, F, Kamakura, T, Komba, S, Nakai, H, Taguchi, H. | | Deposit date: | 2018-10-23 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification, characterization, and structural analyses of a fungal endo-beta-1,2-glucanase reveal a new glycoside hydrolase family.
J.Biol.Chem., 294, 2019
|
|
5LPH
 
 | | Structure of the TOG domain of human Cep104 | | Descriptor: | Centrosomal protein of 104 kDa | | Authors: | van Breugel, M. | | Deposit date: | 2016-08-13 | | Release date: | 2017-01-11 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Ciliopathy-Associated Cep104 Protein Interacts with Tubulin and Nek1 Kinase.
Structure, 25, 2017
|
|
8HBK
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M. | | Deposit date: | 2022-10-29 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
6GM0
 
 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant E279Q | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Duan, J, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8GY6
 
 | |
1I94
 
 | | CRYSTAL STRUCTURES OF THE SMALL RIBOSOMAL SUBUNIT WITH TETRACYCLINE, EDEINE AND IF3 | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Pioletti, M, Schluenzen, F, Harms, J, Zarivach, R, Gluehmann, M, Avila, H, Bartels, H, Jacobi, C, Hartsch, T, Yonath, A, Franceschi, F. | | Deposit date: | 2001-03-18 | | Release date: | 2001-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
EMBO J., 20, 2001
|
|
8H4Y
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HB0
 
 | | Structure of human SGLT2-MAP17 complex with TA1887 | | Descriptor: | (2R,3R,4S,5S,6R)-2-[3-[(4-cyclopropylphenyl)methyl]-4-fluoranyl-indol-1-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5HWA
 
 | | Crystal Structure of MH-K1 chitosanase in substrate-bound form | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Suzuki, M, Saito, A, Ando, A, Miki, K, Saito, J. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1 in complex with chitotetraose
Biomed.Biochim.Acta, 1868, 2024
|
|
6GO6
 
 | | TdT chimera (Loop1 of pol mu) - ternary complex with downstream dsDNA | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*AP*AP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
