5RBW
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library D06b | | Descriptor: | (3-methoxyphenyl)(pyrrolidin-1-yl)methanone, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCC
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library H02b | | Descriptor: | 3-cyclopentyl-1-(piperazin-1-yl)propan-1-one, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCS
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 14 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDC
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 34 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
8WSV
 
 | |
5RDV
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 52 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6OL8
 
 | | Crystal structure of NDM-12 metallo-beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-12, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
2G4O
 
 | | anomalous substructure of 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-isopropylmalate dehydrogenase, CHLORIDE ION, SULFATE ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6T7U
 
 | | Carborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, Carborane inhibitor, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
7BS8
 
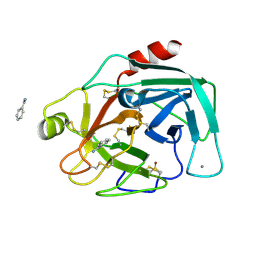 | | Bovine Pancreatic Trypsin with Benzamidine (Cryo) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
5R0W
 
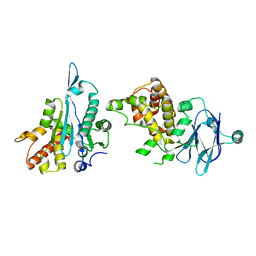 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 10, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1C
 
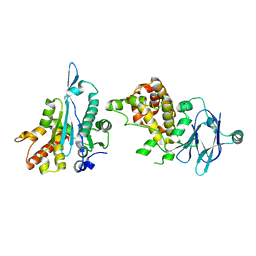 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 27, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5NCG
 
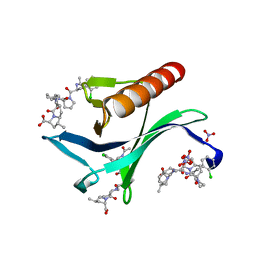 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-9]-OH | | Descriptor: | (3~{S},7~{R},10~{R},11~{R},13~{S})-4-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-11-methyl-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, NITRATE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-04 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5R1S
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 42, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5L8M
 
 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans Bphp collected at LCLS | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Edlund, P, Henry, L, Dods, R, Schmidt, M, Westenhoff, S. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
5R29
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry G09, DMSO-free | | Descriptor: | (2R)-2-(acetylamino)-4-phenylbutanoic acid, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.105 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2R
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 15, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.048 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R38
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 32, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.076 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3N
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 47, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.059 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R16
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 21, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
2G4S
 
 | | Anomalous substructure of NBR1PB1 | | Descriptor: | ACETIC ACID, CHLORIDE ION, Next to BRCA1 gene 1 protein | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5R1K
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 35, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6VWW
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Protein Sci., 29, 2020
|
|
5R1V
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry C04, DMSO-free | | Descriptor: | 2-hydrazinyl-4-methoxypyrimidine, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
2G4W
 
 | | anomalous substructure of ribonuclease A (C2) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
