7FMO
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06D12 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, DIMETHYL SULFOXIDE, N-(2-methoxyphenyl)-2-methyl-L-alanine, ... | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FMU
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06E11 from the F2X-Universal Library | | Descriptor: | (6P)-4-(methylsulfanyl)-6-(2-methyl-1,3-thiazol-4-yl)-1,3,5-triazin-2-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7VH9
 
 | | Solution structure of the chimeric peptide of the first SURP domain of Human SF3A1 and the interacting region of SF1. | | Descriptor: | Splicing factor 3A subunit 1,Splicing factor 1 | | Authors: | Muto, Y, Kuwasako, K, Takizawa, M, Kobayashi, N, Sakamoto, T. | | Deposit date: | 2021-09-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction between the first SURP domain of the SF3A1 subunit in U2 snRNP and the human splicing factor SF1.
Protein Sci., 31, 2022
|
|
7FN2
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06G08 from the F2X-Universal Library | | Descriptor: | 1-[2-(morpholin-4-yl)pyridin-4-yl]methanamine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FN7
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06H08 from the F2X-Universal Library | | Descriptor: | 6-chloro-N-(3-methoxypropyl)pyrimidin-4-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
1QKI
 
 | | X-RAY STRUCTURE OF HUMAN GLUCOSE 6-PHOSPHATE DEHYDROGENASE (VARIANT CANTON R459L) COMPLEXED WITH STRUCTURAL NADP+ | | Descriptor: | GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL, GLYCOLIC ACID, ... | | Authors: | Au, S.W.N, Gover, S, Lam, V.M.S, Adams, M.J. | | Deposit date: | 1999-07-20 | | Release date: | 2000-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human Glucose-6-Phosphate Dehydrogenase: The Crystal Structure Reveals a Structural Nadp+ Molecule and Provides Insights Into Enzyme Deficiency
Structure, 8, 2000
|
|
7FNB
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07A12 from the F2X-Universal Library | | Descriptor: | 2-cyano-N-methyl-N-phenylacetamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
6MD9
 
 | |
7FNL
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07D01 from the F2X-Universal Library | | Descriptor: | 1-ethyl-N-[(thiophen-2-yl)methyl]-1H-tetrazol-5-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FN0
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06G05 from the F2X-Universal Library | | Descriptor: | 4-{[(propan-2-yl)oxy]methyl}benzoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FN5
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06H04 from the F2X-Universal Library | | Descriptor: | 5-bromo-N-propylthiophene-3-carboxamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3BZV
 
 | | Crystal structural of the mutated T264A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
7FNG
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07B12 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-(2-AMINOETHYL)-P-CHLOROBENZAMIDE, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3BT1
 
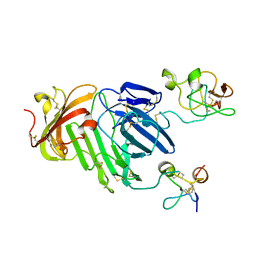 | | Structure of urokinase receptor, urokinase and vitronectin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator, ... | | Authors: | Huang, M. | | Deposit date: | 2007-12-27 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes
Nat.Struct.Mol.Biol., 15, 2008
|
|
7FNS
 
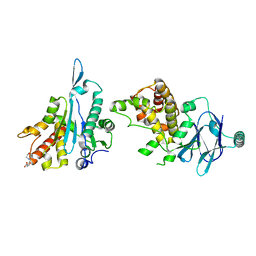 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07E03 from the F2X-Universal Library | | Descriptor: | 2-[(morpholin-4-yl)methyl]furan-3-carboxylic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FO2
 
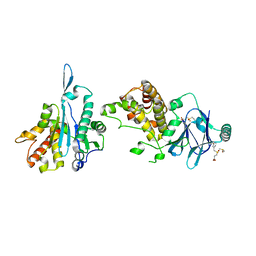 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07G04 from the F2X-Universal Library | | Descriptor: | 3-[(3-bromophenyl)methanesulfonyl]propanoic acid, A1 cistron-splicing factor AAR2, DIMETHYL SULFOXIDE, ... | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOB
 
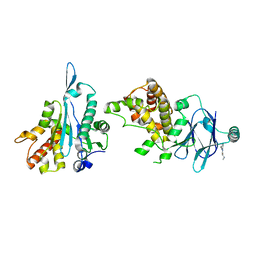 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07H11 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-(3-fluorophenyl)-N~2~-methylglycinamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOI
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08B06 from the F2X-Universal Library | | Descriptor: | 3-fluoro-N-[2-(1H-imidazol-1-yl)ethyl]benzamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3C05
 
 | | Crystal structure of Acostatin from Agkistrodon Contortrix Contortrix | | Descriptor: | Disintegrin acostatin alpha, Disintegrin acostatin-beta, SULFATE ION | | Authors: | Moiseeva, N, Bau, R, Allaire, M. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of acostatin, a dimeric disintegrin from southern copperhead
(Agkistrodon contortrix contortrix) at 1.7 A resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7BGS
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7FNM
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07D05 from the F2X-Universal Library | | Descriptor: | 5-bromo-N-(2-methoxyethyl)pyridine-3-carboxamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FNU
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07F03 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, methyl [(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FNY
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07F10 from the F2X-Universal Library | | Descriptor: | 4-[(acetyloxy)methyl]benzoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3F1Y
 
 | | Mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mannosyl-3-phosphoglycerate synthase | | Authors: | Macedo-Ribeiro, S, Pereira, P.J.B, Empadinhas, N, da Costa, M.S. | | Deposit date: | 2008-10-28 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characterization of a novel mannosyl-3-phosphoglycerate synthase from Rubrobacter xylanophilus reveals its dual substrate specificity
Mol.Microbiol., 79, 2011
|
|
7FOP
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08C08 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-[(1S)-1-cyanopropyl]benzamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
