3PP6
 
 | | REP1-NXSQ fatty acid transporter Y128F mutant | | Descriptor: | PALMITOLEIC ACID, ReP1-NCXSQ | | Authors: | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | Deposit date: | 2010-11-24 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7WWV
 
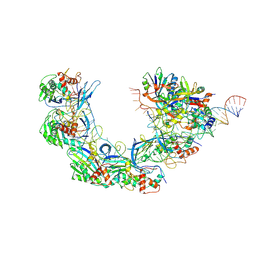 | | DNA bound-ICP1 Csy complex | | Descriptor: | Csy1, Csy2, Csy3, ... | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WWU
 
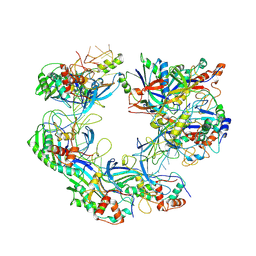 | | ICP1 Csy complex | | Descriptor: | Csy1, Csy2, Csy3, ... | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1EJ2
 
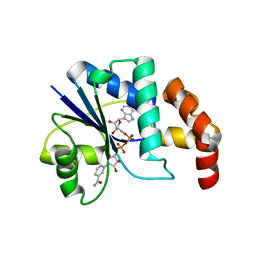 | | Crystal structure of methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase with bound NAD+ | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F, Midwest Center for Structural Genomics (MCSG), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-29 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
1EL8
 
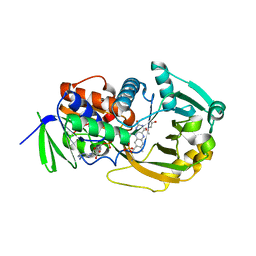 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYLSELENO]CETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
1EVX
 
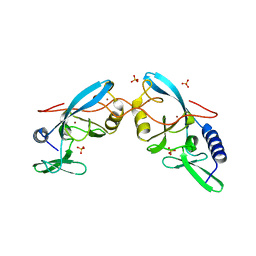 | | APO CRYSTAL STRUCTURE OF THE HOMING ENDONUCLEASE, I-PPOI | | Descriptor: | INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SULFATE ION, ZINC ION | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
3PPT
 
 | | REP1-NXSQ fatty acid transporter | | Descriptor: | PALMITOLEIC ACID, ReP1-NCXSQ | | Authors: | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | Deposit date: | 2010-11-25 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1EKJ
 
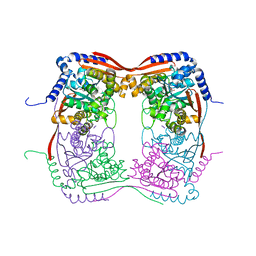 | |
1EWI
 
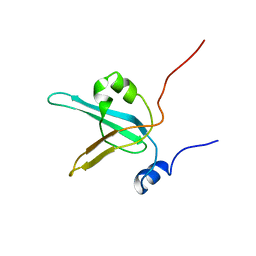 | | HUMAN REPLICATION PROTEIN A: GLOBAL FOLD OF THE N-TERMINAL RPA-70 DOMAIN REVEALS A BASIC CLEFT AND FLEXIBLE C-TERMINAL LINKER | | Descriptor: | REPLICATION PROTEIN A | | Authors: | Jacobs, D.M, Lipton, A.S, Isern, N.G, Daughdrill, G.W, Lowry, D.F, Gomes, X, Wold, M.S. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker.
J.Biomol.NMR, 14, 1999
|
|
1EGL
 
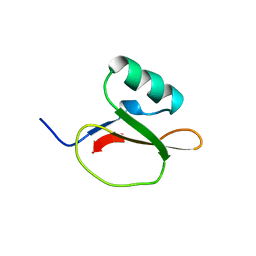 | |
1EL5
 
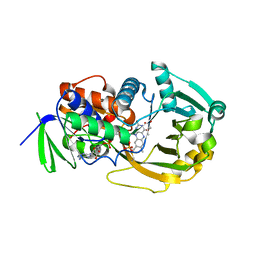 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR DIMETHYLGLYCINE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, N,N-DIMETHYLGLYCINE, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
1EA6
 
 | |
1E7M
 
 | |
1E9W
 
 | | Structure of the macrocycle thiostrepton solved using the anomalous dispersive contribution from sulfur | | Descriptor: | THIOSTREPTON | | Authors: | Bond, C.S, Shaw, M.P, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2000-10-27 | | Release date: | 2001-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the Macrocycle Thiostrepton Solved Using the Anomalous Dispersive Contribution from Sulfur
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7Y8O
 
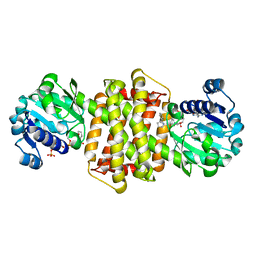 | | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SciR | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole
to be published
|
|
1F1W
 
 | | SRC SH2 THREF1TRP MUTANT COMPLEXED WITH THE PHOSPHOPEPTIDE S(PTR)VNVQN | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, S(PTR)VNVQN PHOSPHOPEPTIDE | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-20 | | Release date: | 2000-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3RDO
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin | | Descriptor: | BIOTIN, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE5
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
7X5C
 
 | | Solution structure of Tetrahymena p75OB1-p50PBM | | Descriptor: | Telomerase associated protein p50PBM, Telomerase-associated protein p75OB1 | | Authors: | Wu, B, Tang, T, Xue, H.J, Wu, J, Lei, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Association of the CST complex and p50 in Tetrahymena is crucial for telomere maintenance
Structure, 2022
|
|
3RDS
 
 | | Crystal structure of the refolded R7-2 streptavidin | | Descriptor: | PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
1F2F
 
 | | SRC SH2 THREF1TRP MUTANT | | Descriptor: | PHOSPHATE ION, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
7WKP
 
 | | ICP1 Csy4 | | Descriptor: | 3' stem loop crRNA, Csy4 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WKO
 
 | | ICP1 Csy1-2 complex | | Descriptor: | Csy1, Csy2 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
