1URY
 
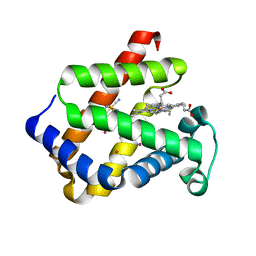 | | cytoglobin cavities | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cytoglobin Cavities
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1V3Z
 
 | |
1VB0
 
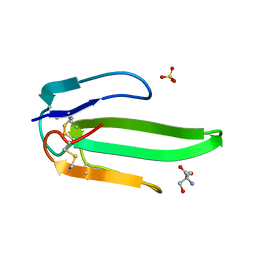 | | Atomic resolution structure of atratoxin-b, one short-chain neurotoxin from Naja atra | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Liu, Q, Teng, M, Niu, L, Huang, Q, Hao, Q. | | Deposit date: | 2004-02-20 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
1VB7
 
 | |
1V4S
 
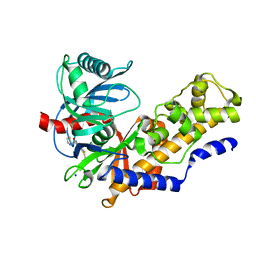 | | Crystal structure of human glucokinase | | Descriptor: | 2-AMINO-4-FLUORO-5-[(1-METHYL-1H-IMIDAZOL-2-YL)SULFANYL]-N-(1,3-THIAZOL-2-YL)BENZAMIDE, SODIUM ION, alpha-D-glucopyranose, ... | | Authors: | Kamata, K, Mitsuya, M, Nishimura, T, Eiki, J, Nagata, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase
Structure, 12, 2004
|
|
1V5R
 
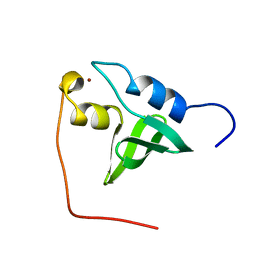 | | Solution Structure of the Gas2 Domain of the Growth Arrest Specific 2 Protein | | Descriptor: | Growth-arrest-specific protein 2, ZINC ION | | Authors: | Miyamoto, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2005-01-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Gas2 Domain of the Growth Arrest Specific 2 Protein
To be Published
|
|
1V64
 
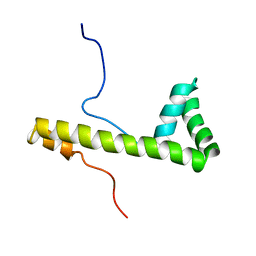 | | Solution structure of the 3rd HMG box of mouse UBF1 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 3rd HMG box of mouse UBF1
To be Published
|
|
1UUU
 
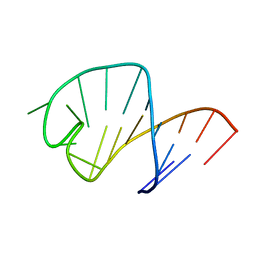 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
1V82
 
 | | Crystal structure of human GlcAT-P apo form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V8B
 
 | | Crystal structure of a hydrolase | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, adenosylhomocysteinase | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Shiraiwa, K, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-01-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of S-Adenosyl-l-Homocysteine Hydrolase from the Human Malaria Parasite Plasmodium falciparum
J.Mol.Biol., 343, 2004
|
|
1V9W
 
 | | Solution structure of mouse putative 42-9-9 protein | | Descriptor: | putative 42-9-9 protein | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse putative 42-9-9 protein
To be Published
|
|
4YTN
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[3-(pentafluorophenoxy)phenyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New Insights into the Design of Inhibitors Targeted for Parasitic Anaerobic Energy Metabolism
To Be Published
|
|
1UR4
 
 | | The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GALACTANASE, ... | | Authors: | Ryttersgaard, C, Le Nours, J, Lo Leggio, L, Jorgensen, C.T, Christensen, L.L.H, Bjornvad, M, Larsen, S. | | Deposit date: | 2003-10-24 | | Release date: | 2004-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Endo-Beta-1,4-Galactanase from Bacillus Licheniformis in Complex with Two Oligosaccharide Products
J.Mol.Biol., 341, 2004
|
|
1UU5
 
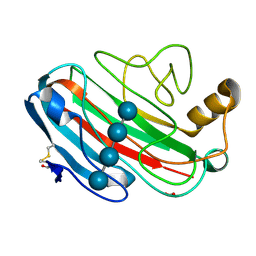 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A SOAKED WITH CELLOTETRAOSE | | Descriptor: | ACETATE ION, ENDO-BETA-1,4-GLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2003-12-15 | | Release date: | 2004-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
1V29
 
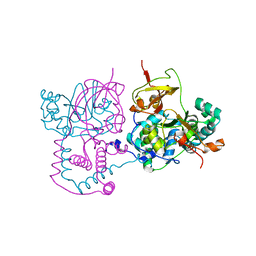 | | Crystal structure of Nitrile hydratase from a thermophile Bacillus smithii | | Descriptor: | COBALT (II) ION, nitrile hydratase a chain, nitrile hydratase b chain | | Authors: | Hourai, S, Miki, M, Takashima, Y, Mitsuda, S, Yanagi, K. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of nitrile hydratase from a thermophilic Bacillus smithii
Biochem.Biophys.Res.Commun., 312, 2003
|
|
1V43
 
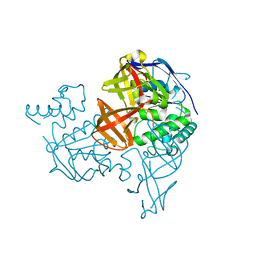 | | Crystal Structure of ATPase subunit of ABC Sugar Transporter | | Descriptor: | sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-11-08 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
1V53
 
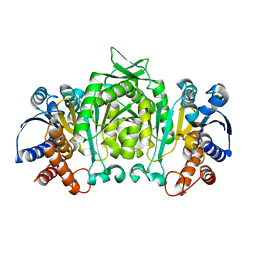 | | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans | | Descriptor: | 3-isopropylmalate dehydrogenase | | Authors: | Fujita, K, Minami, H, Suzuki, K, Tsunoda, M, Sekiguchi, T, Mizui, R, Tsuzaki, S, Nakamura, S, Takenaka, A. | | Deposit date: | 2003-11-20 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of 3-isopropylmalate dehydrogenase from Bacillus coagulans
To be Published
|
|
3COU
 
 | | Crystal structure of human Nudix motif 16 (NUDT16) | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 16 | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Van Den Berg, S, Welin, M, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Nudix motif 16 (NUDT16).
To be Published
|
|
1V30
 
 | | Crystal Structure Of Uncharacterized Protein PH0828 From Pyrococcus horikoshii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hypothetical UPF0131 protein PH0828 | | Authors: | Tajika, Y, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-10-21 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of hypothetical protein PH0828 from Pyrococcus horikoshii.
Proteins, 57, 2004
|
|
1V46
 
 | |
1V5P
 
 | | Solution Structure of the N-terminal Pleckstrin Homology Domain Of TAPP2 from Mouse | | Descriptor: | pleckstrin homology domain-containing, family A | | Authors: | Li, H, Hayashi, F, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Pleckstrin Homology Domain Of TAPP2 from Mouse
To be Published
|
|
1V6E
 
 | | Solution Structure of a N-terminal Ubiquitin-like Domain in Mouse Tubulin-specific Chaperone B | | Descriptor: | cytoskeleton-associated protein 1 | | Authors: | Zhao, C, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a N-terminal Ubiquitin-like Domain in Mouse Tubulin-specific Chaperone B
To be Published
|
|
1V76
 
 | |
1V5K
 
 | | Solution structure of the CH domain from mouse EB-1 | | Descriptor: | microtubule-associated protein, RP/EB family, member 1 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from mouse EB-1
To be Published
|
|
1V62
 
 | | Solution structure of the 3rd PDZ domain of GRIP2 | | Descriptor: | KIAA1719 protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 3rd PDZ domain of GRIP2
To be Published
|
|
