3MDD
 
 | |
6F7M
 
 | | NMR structure of EphA2-Sam stapled peptides (S13ST) | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6F7N
 
 | | NMR structure of EphA2-Sam stapled peptides (S13STshort) | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
4KYH
 
 | | Crystal structure of mouse glyoxalase I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
6F7O
 
 | | NMR structure of an Odin-Sam1 stapled peptide | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 1A | | Authors: | Leone, M, Mercurio, F.A. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6HHE
 
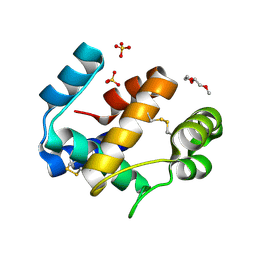 | | Crystal structure of the medfly Odorant Binding Protein CcapOBP22/CcapOBP69a | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Odorant binding protein OBP69a, SULFATE ION | | Authors: | Falchetto, M, Ciossani, G, Nenci, S, Mattevi, A, Gasperi, G, Forneris, F. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication.
Insect Mol.Biol., 28, 2019
|
|
4KYO
 
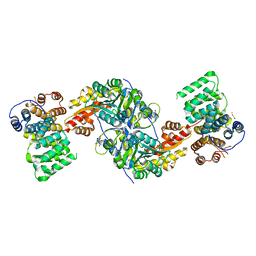 | | Alanine-glyoxylate aminotransferase variant K390A in complex with the TPR domain of human Pex5p | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, ... | | Authors: | Fodor, K, Lou, Y, Wilmanns, M. | | Deposit date: | 2013-05-29 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
4ZZB
 
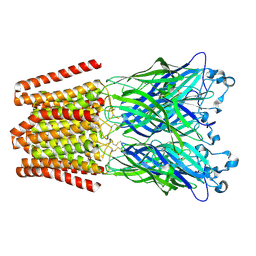 | | The GLIC pentameric Ligand-Gated Ion Channel Locally-closed form complexed to xenon | | Descriptor: | ACETATE ION, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Fourati, Z, Prange, T, Delarue, M, Colloc'h, N. | | Deposit date: | 2015-05-22 | | Release date: | 2016-03-02 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Xenon Inhibition in a Cationic Pentameric Ligand-Gated Ion Channel.
Plos One, 11, 2016
|
|
7ZNY
 
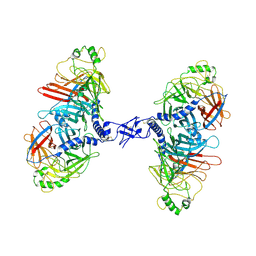 | | Cryo-EM structure of the canine distemper virus tetrameric attachment glycoprotein | | Descriptor: | Hemagglutinin glycoprotein | | Authors: | Kalbermatter, D, Jeckelmann, J.-M, Wyss, M, Plattet, P, Fotiadis, D. | | Deposit date: | 2022-04-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and supramolecular organization of the canine distemper virus attachment glycoprotein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4L0K
 
 | |
1LJL
 
 | | Wild Type pI258 S. aureus arsenate reductase | | Descriptor: | POTASSIUM ION, arsenate reductase | | Authors: | Messens, J, Martins, J.C, Van Belle, K, Brosens, E, Desmyter, A, De Gieter, M, Wieruszeski, J.M, Willem, R, Wyns, L, Zegers, I. | | Deposit date: | 2002-04-21 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3MGK
 
 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
5A0P
 
 | | Apo-structure of metalloprotease Zmp1 from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
4L33
 
 | | Tankyrase 2 in complex with cyanomethyl 4-(4-oxo-4H-chromen-2-yl)benzoate | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2013-06-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity.
J.Med.Chem., 56, 2013
|
|
5A0X
 
 | | Substrate peptide-bound structure of metalloprotease Zmp1 variant E143AY178F from Clostridium difficile | | Descriptor: | SUBSTRATE PEPTIDE, ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
3IHL
 
 | | Human CTPS2 crystal structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CTP synthase 2, PHOSPHATE ION | | Authors: | Moche, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Kragh Nielsen, T, Nyman, T, Persson, C, Roos, A.K, Sagemark, J, Schueler, H, Schutz, P, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human CTPS2 crystal structure
To be Published
|
|
1LO5
 
 | | Crystal structure of the D227A variant of Staphylococcal enterotoxin A in complex with human MHC class II | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DR-1 beta chain, ... | | Authors: | Petersson, K, Thunnissen, M, Forsberg, G, Walse, B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a SEA Variant in Complex with MHC Class II Reveals the Ability of SEA to Crosslink MHC Molecules
Structure, 10, 2002
|
|
6FRP
 
 | |
6HJA
 
 | | Xray structure of GLIC in complex with glutarate | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECANE, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the binding of monocarboxylates and dicarboxylates at pharmacologically relevant extracellular sites of a pentameric ligand-gated ion channel.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5A16
 
 | | Crystal structure of Fab4201 raised against Human Erythrocyte Anion Exchanger 1 | | Descriptor: | FAB4201 HEAVY CHAIN | | Authors: | Arakawa, T, Kobayashi-Yugiri, T, Alguel, Y, Weyand, S, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Ikeda-Suno, C, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Anion Exchanger Domain of Human Erythrocyte Band 3
Science, 350, 2015
|
|
5A8P
 
 | | Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide B | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
4L7C
 
 | | Structure of keap1 kelch domain with 2-{[(1S)-2-{[(1R,2S)-2-(1H-tetrazol-5-yl)cyclohexyl]carbonyl}-1,2,3,4-tetrahydroisoquinolin-1-yl]methyl}-1H-isoindole-1,3(2H)-dione | | Descriptor: | 2-{[(1S)-2-{[(1R,2S)-2-(1H-tetrazol-5-yl)cyclohexyl]carbonyl}-1,2,3,4-tetrahydroisoquinolin-1-yl]methyl}-1H-isoindole-1,3(2H)-dione, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
6FLS
 
 | |
1LPU
 
 | | Low Temperature Crystal Structure of the Apo-form of the catalytic subunit of protein kinase CK2 from Zea mays | | Descriptor: | BENZAMIDINE, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
6HM1
 
 | | Structural and thermodynamic signatures of ligand binding to an enigmatic chitinase-D from Serratia proteamaculans | | Descriptor: | 1,2-ETHANEDIOL, ALLOSAMIDIN, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Dalhus, B, Vaaje-Kolstad, G, Eijsink, V.G.H, Sorlie, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and Thermodynamic Signatures of Ligand Binding to the Enigmatic Chitinase D of Serratia proteamaculans.
J.Phys.Chem.B, 123, 2019
|
|
