5LSO
 
 | |
5LTH
 
 | |
5AUW
 
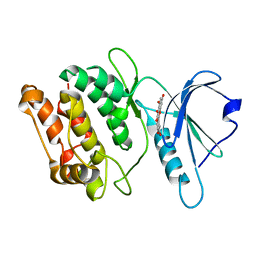 | | Crystal structure of DAPK1 in complex with quercetin. | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Death-associated protein kinase 1 | | Authors: | Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2015-06-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into the Interactions between Death-Associated Protein Kinase 1 and Natural Flavonoids.
J.Med.Chem., 58, 2015
|
|
7C6F
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in an open state | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
3OZR
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, bisubstrate inhibitor, no substituent in the adenine site - humanized form | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
7C6M
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotetraose (Form I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5BSE
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, Pyrroline-5-carboxylate reductase | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
7CCU
 
 | |
5LXX
 
 | | High-resolution structure of human collapsin response mediator protein 2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydropyrimidinase-related protein 2, SULFATE ION | | Authors: | Myllykoski, M, Hensley, K, Kursula, P. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Collapsin response mediator protein 2: high-resolution crystal structure sheds light on small-molecule binding, post-translational modifications, and conformational flexibility.
Amino Acids, 49, 2017
|
|
5LP2
 
 | |
5LPF
 
 | | Kallikrein-related peptidase 10 | | Descriptor: | Kallikrein-10, SULFATE ION | | Authors: | Goettig, P, Debela, M, Magdolen, V, Bode, W, Brandstetter, H. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Zn2+ inhibition of the zymogen-like kallikrein-related peptidase 10.
Biol.Chem., 397, 2016
|
|
5B1N
 
 | | DHp domain structure of EnvZ from Escherichia coli | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
5AYH
 
 | |
5AYT
 
 | | Crystal structure of transthyretin in complex with L6 | | Descriptor: | 2-oxidanyl-6-(phenylcarbonyl)benzo[de]isoquinoline-1,3-dione, Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Kai, H. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural stabilization of transthyretin by a new compound, 6-benzoyl-2-hydroxy-1H-benzo[de]isoquinoline-1,3(2H)-dione
J. Pharmacol. Sci., 129, 2015
|
|
5B22
 
 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
5LU7
 
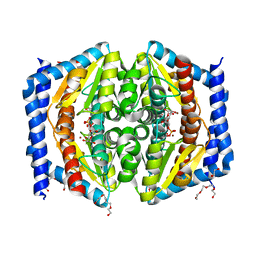 | | Heptose isomerase GmhA mutant - D61A | | Descriptor: | 1,2-ETHANEDIOL, 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, Phosphoheptose isomerase, ... | | Authors: | Vivoli, M, Harmer, N.J, Pang, J. | | Deposit date: | 2016-09-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
5AZ1
 
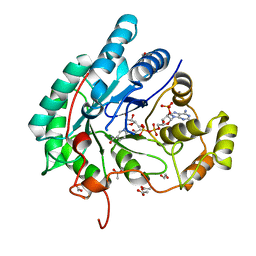 | | Crystal structure of aldo-keto reductase (AKR2E5) complexed with NADPH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yamamoto, K, Higashiura, A, Suzuki, M, Nakagawa, A. | | Deposit date: | 2015-09-15 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5B2K
 
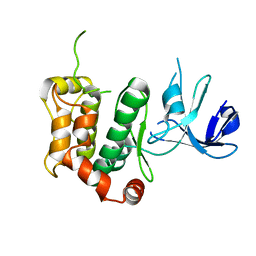 | | A crucial role of Cys218 in the stabilization of an unprecedented auto-inhibition form of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, T, Matsumoto, T, Kirii, Y, Sawa, M, Kinoshita, T. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A crucial role of Cys218 in configuring an unprecedented auto-inhibition form of MAP2K7
Biochem.Biophys.Res.Commun., 473, 2016
|
|
2LLJ
 
 | | Structure of a bis-naphthalene bound to a thymine-thymine DNA mismatch | | Descriptor: | 6,22-dioxa-3,9,19,25-tetraazoniapentacyclo[25.5.3.3~11,17~.0~14,37~.0~30,34~]octatriaconta-1(33),11(38),12,14(37),15,17(36),27,29,31,34-decaene, DNA (5'-D(*CP*GP*TP*CP*GP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*CP*TP*TP*CP*GP*AP*CP*G)-3') | | Authors: | Jourdan, M, Granzhan, A, Guillot, R, Dumy, P, Teulade-Fichou, M. | | Deposit date: | 2011-11-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Double threading through DNA: NMR structural study of a bis-naphthalene macrocycle bound to a thymine-thymine mismatch.
Nucleic Acids Res., 40, 2012
|
|
7CJG
 
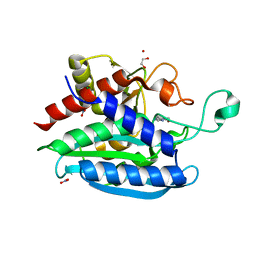 | | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | 5,6-DIMETHYLBENZIMIDAZOLE, GLYCEROL, Glutamine cyclotransferase-related protein, ... | | Authors: | Ruiz-Carrillo, D, Lamers, S, Feng, Q, Yu, S, Sun, B, Jiang, J, Lukman, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase.
Biol.Chem., 402, 2021
|
|
5M2A
 
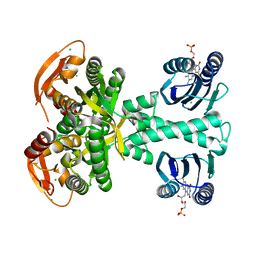 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-10-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
1TJ1
 
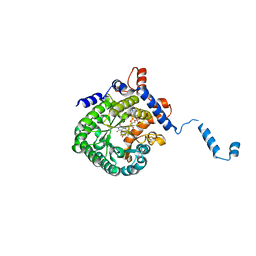 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
5B0U
 
 | |
5B1J
 
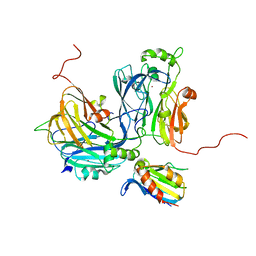 | | Crystal structure of the electron-transfer complex of copper nitrite reductase with a cupredoxin | | Descriptor: | Blue copper protein, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Nojiri, M, Koteishi, H, Yoneda, R, Hira, D. | | Deposit date: | 2015-12-04 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Function of Copper Nitrite Reductase
Metalloenzymes in denitrification: Applications and Environmental impacts, 2016
|
|
5LZ3
 
 | |
