8ZMG
 
 | | Crystal structure of an inverse agonist antipsychotic drug pimavanserin-bound 5-HT2A | | Descriptor: | 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, Pimavanserin | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
5OXU
 
 | |
2B60
 
 | | Structure of HIV-1 protease mutant bound to Ritonavir | | Descriptor: | GLYCEROL, Gag-Pol polyprotein, RITONAVIR | | Authors: | Clemente, J.C, Stow, L.R, Janka, L.K, Jeung, J.A, Desai, K.A, Govindasamy, L, Agbandje-McKenna, M, McKenna, R, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivo, kinetic, and structural analysis of the development of ritonavir resistance
To be Published
|
|
2AHE
 
 | | Crystal structure of a soluble form of CLIC4. intercellular chloride ion channel | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Littler, D.R, Assaad, N.N, Harrop, S.J, Brown, L.J, Pankhurst, G.J, Luciani, P, Aguilar, M.-I, Mazzanti, M, Berryman, M.A, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4.
FEBS J., 272, 2005
|
|
2AHV
 
 | | Crystal Structure of Acyl-CoA transferase from E. coli O157:H7 (YdiF)-thioester complex with CoA- 1 | | Descriptor: | COENZYME A, putative enzyme YdiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
9B44
 
 | |
3POR
 
 | |
9AUR
 
 | | Crystal structure of loop-closed A21 2'-OMe dumbbell RNA bridged by glycine | | Descriptor: | Fab BL3-6 heavy chain, Fab BL3-6 light chain, GLYCINE, ... | | Authors: | Radakovic, A, Lewicka, A, Todisco, M, Aitken, H.R.M, Weiss, Z, Kim, S, Bannan, A, Piccirilli, J.A, Szostak, J.W. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A potential role for RNA aminoacylation prior to its role in peptide synthesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9AUS
 
 | | Crystal structure of loop-closed dumbbell RNA bridged by glycine | | Descriptor: | Fab BL3-6 heavy chain, Fab BL3-6 light chain, GLYCINE, ... | | Authors: | Radakovic, A, Lewicka, A, Todisco, M, Aitken, H.R.M, Weiss, Z, Kim, S, Bannan, A, Piccirilli, J.A, Szostak, J.W. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A potential role for RNA aminoacylation prior to its role in peptide synthesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8YQ9
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S) complexed with FB6, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[4-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxypropoxy]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D, Saeyang, T, Arwon, U, Hoarau, M, Decharuangsilp, S, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel flexible biphenyl Pf DHFR inhibitors with improved antimalarial activity.
Rsc Med Chem, 15, 2024
|
|
8YQ8
 
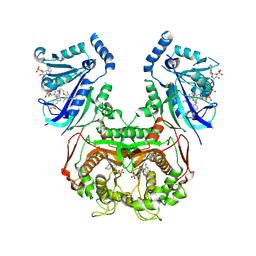 | | Quadruple mutant (N51I+C59R+S108N+I164L) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S) complexed with FB8, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-[4-[4-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxybutoxy]phenyl]benzoic acid, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D, Saeyang, T, Arwon, U, Hoarau, M, Decharuangsilp, S, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel flexible biphenyl Pf DHFR inhibitors with improved antimalarial activity.
Rsc Med Chem, 15, 2024
|
|
9AYM
 
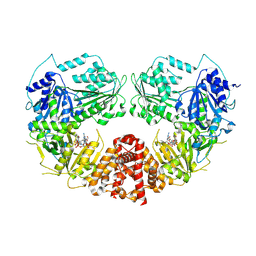 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-acetone-CoA bisubstrate probe | | Descriptor: | RNA cytidine acetyltransferase, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-[2-[3-[1-[(2~{R},3~{R},4~{R},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]sulfanyl-2-oxidanylidene-propyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
9B0I
 
 | |
6LOO
 
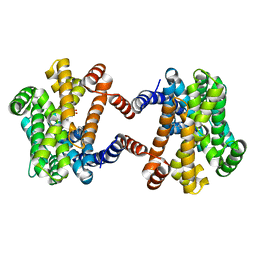 | | Crystal Structure of Class IB terpene synthase bound with geranylcitronellyl diphosphate | | Descriptor: | Tetraprenyl-beta-curcumene synthase, phosphono [(3~{R},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-6,10,14-trienyl] hydrogen phosphate, phosphono [(3~{S},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-6,10,14-trienyl] hydrogen phosphate | | Authors: | Fujihashi, M, Inagi, H, Miki, K. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of Class IB Terpene Synthase: The First Crystal Structure Bound with a Substrate Surrogate.
Acs Chem.Biol., 15, 2020
|
|
6LU3
 
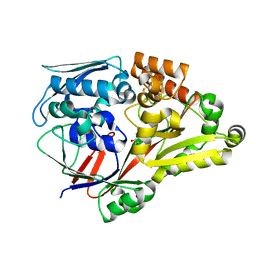 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans complexed with 4-hydroxybenzoate hydrazide | | Descriptor: | 4-oxidanylbenzohydrazide, Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
5OVL
 
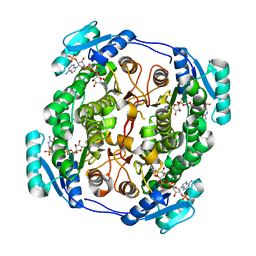 | | crystal structure of MabA bound to NADP+ from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OXT
 
 | |
6MGC
 
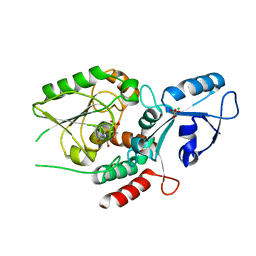 | | Escherichia coli KpsC, N-terminal domain | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsule polysaccharide export protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
6LU4
 
 | | Crystal structure of the substrate binding protein from Microbacterium hydrocarbonoxydans complexed with propylparaben | | Descriptor: | Substrate binding protein, propyl 4-hydroxybenzoate | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
4DPV
 
 | | PARVOVIRUS/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*CP*TP*CP*TP*TP*GP*C)-3'), MAGNESIUM ION, PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Chapman, M.S, Rossmann, M.G. | | Deposit date: | 1996-02-01 | | Release date: | 1997-04-01 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Canine parvovirus capsid structure, analyzed at 2.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
4MG6
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with benzylbutylphtalate | | Descriptor: | Estrogen receptor, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
5WSU
 
 | | Crystal structure of Myosin VIIa IQ5-SAH in complex with apo-CaM | | Descriptor: | Calmodulin, Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
4LT4
 
 | | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution
To be Published
|
|
3ZLW
 
 | | Crystal structure of MEK1 in complex with fragment 3 | | Descriptor: | (1R)-1-hydroxy-1-methyl-2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-5-one, DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 1 MAPK/ERK KINASE 1, MEK 1, ... | | Authors: | Amaning, K, Lowinsky, M, Vallee, F, Steier, V, Marcireau, C, Ugolini, A, Delorme, C, McCort, G, Andouche, C, Vougier, S, Llopart, S, Halland, N, Rak, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Use of Virtual Screening and Differential Scanning Fluorimetry for the Rapid Identification of Fragments Active Against Mek1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
