8Z5L
 
 | | Crystal structure of metallo-beta-lactamse, IMP-1, complexed with a quinolinone-based inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-cyclohexylethyl-[3-(4-methylphenoxy)propyl]amino]phenyl]propanoic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Nimura, S, Guo, Y, Kondo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of Inhibitory Compounds for Metallo-beta-lactamase through Computational Design and Crystallographic Analysis.
Biochemistry, 63, 2024
|
|
7QUX
 
 | | Crystal structure of P7C8 bound to CK2alpha | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Atkinson, E, Iegre, J, Brear, P, Baker, D, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2022-01-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Development of small cyclic peptides targeting the CK2 alpha / beta interface.
Chem.Commun.(Camb.), 58, 2022
|
|
4M3P
 
 | | Betaine-Homocysteine S-Methyltransferase from Homo sapiens complexed with Homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Betaine--homocysteine S-methyltransferase 1, POTASSIUM ION, ... | | Authors: | Koutmos, M, Yamada, K, Mladkova, J, Paterova, J, Diamond, C.E, Tryon, K, Jungwirth, P, Garrow, T.A, Jiracek, J. | | Deposit date: | 2013-08-06 | | Release date: | 2014-06-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Specific potassium ion interactions facilitate homocysteine binding to betaine-homocysteine S-methyltransferase.
Proteins, 82, 2014
|
|
4MHR
 
 | | Crystal structure of EctD from S. alaskensis in its apoform | | Descriptor: | Ectoine hydroxylase | | Authors: | Widderich, N, Hoeppner, A, Pittelkow, M, Heider, J, Smits, S.H, Bremer, E. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
5J81
 
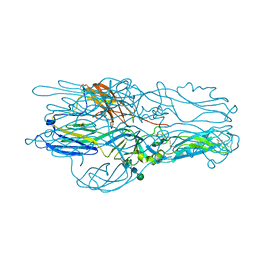 | | Crystal structure of Glycoprotein C from Puumala virus in the post-fusion conformation (pH 6.0) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETATE ION, Envelopment polyprotein, ... | | Authors: | Willensky, S, Dessau, M. | | Deposit date: | 2016-04-07 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Glycoprotein C from a Hantavirus in the Post-fusion Conformation.
Plos Pathog., 12, 2016
|
|
3ZRX
 
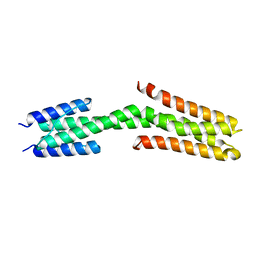 | |
5JES
 
 | | Human carbonic anhydrase II (V121T) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.205 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7QNV
 
 | |
7QOB
 
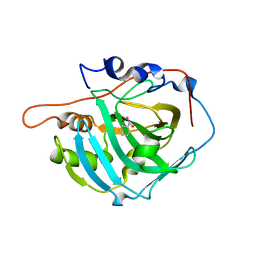 | |
5JN2
 
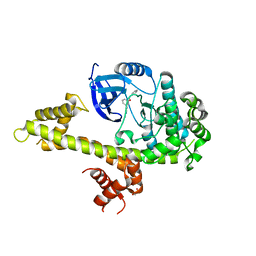 | | Crystal structure of TgCDPK1 bound to NVPACU106 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-domain protein kinase 1, ... | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lovato, D.V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TgCDPK1 bound to NVPACU106
To Be Published
|
|
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | Descriptor: | BARIUM ION, TIP60 K67E mutant | | Authors: | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | Deposit date: | 2022-04-24 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
5ITR
 
 | | Crystal Structure of Human NEIL1(P2G) bound to duplex DNA containing THF | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7QVK
 
 | | NM-02 in complex with HER2-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NM-02, Receptor tyrosine-protein kinase erbB-2, ... | | Authors: | Cowan, R, Hall, G, Carr, M. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Co-crystallisation and humanisation of an anti-HER2 single-domain antibody as a theranostic tool.
Plos One, 18, 2023
|
|
3ZX0
 
 | | NTPDase1 in complex with Heptamolybdate | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for a domain motion in rat nucleoside triphosphate diphosphohydrolase (NTPDase) 1.
J. Mol. Biol., 415, 2012
|
|
5IY4
 
 | |
5IZN
 
 | | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6 | | Descriptor: | 50S ribosomal protein L25, PHOSPHATE ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-25 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6
To Be Published
|
|
3ZRW
 
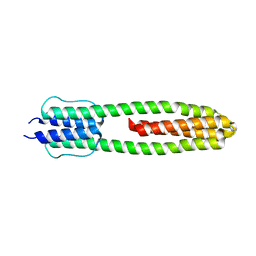 | | The structure of the dimeric Hamp-Dhp fusion A291V mutant | | Descriptor: | AF1503 PROTEIN, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J, Lupas, A.N. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
5J02
 
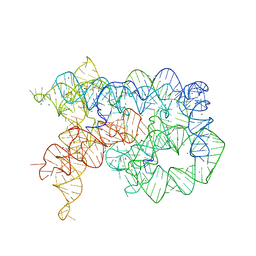 | | Structure of the lariat form of a chimeric derivative of the Oceanobacillus iheyensis group II intron in the presence of NH4+, MG2+ and an inactive 5' exon. | | Descriptor: | 5' EXON ANALOG (5'-R(*CP*UP*GP*UP*UP*AP*(5MU))-3'), AMMONIUM ION, GROUP II INTRON LARIAT, ... | | Authors: | Costa, M, Walbott, H, Monachello, D, Westhof, E, Michel, F. | | Deposit date: | 2016-03-26 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Crystal structures of a group II intron lariat primed for reverse splicing.
Science, 354, 2016
|
|
5J2W
 
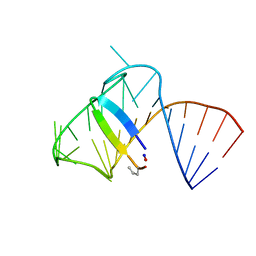 | | Intermediate state lying on the pathway of release of Tat from HIV-1 TAR. | | Descriptor: | Apical region (29mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7Q4H
 
 | |
7Q4J
 
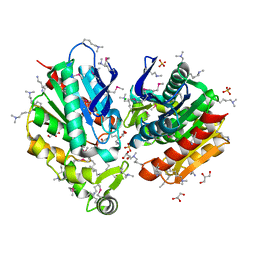 | |
8ZMU
 
 | |
3ZR4
 
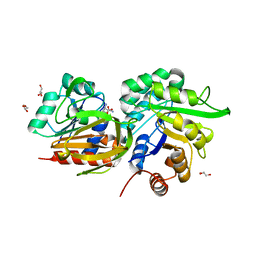 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | Descriptor: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | Authors: | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2011-06-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
7QAC
 
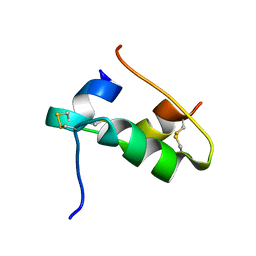 | | The T2 structure of polycrystalline cubic human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Karavassili, F, Triandafillidis, D.P, Valmas, A, Spiliopoulou, M, Fili, S, Kontou, P, Bowler, M.W, Von Dreele, R.B, Fitch, A, Margiolaki, I. | | Deposit date: | 2021-11-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | POWDER DIFFRACTION (2.29 Å) | | Cite: | The T 2 structure of polycrystalline cubic human insulin.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5JEG
 
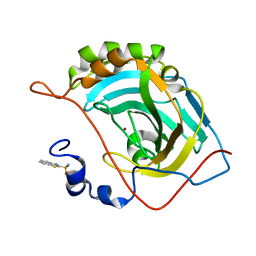 | | Human carbonic anhydrase II (V121I) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
