2J9N
 
 | | Robotically harvested Trypsin complexed with Benzamidine containing polypeptide mediated crystal contacts | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Viola, R, Carman, P, Walsh, J, Miller, E, Benning, M, Frankel, D, McPherson, A, Cudney, R, Rupp, B. | | Deposit date: | 2006-11-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Operator Assisted Harvesting of Protein Crystals Using a Universal Micromanipulation Robot.
J.Appl.Crystallogr., 40, 2007
|
|
4XVP
 
 | | X-ray structure of bGFP-C / EGFP complex | | Descriptor: | BGFP-C, Green fluorescent protein | | Authors: | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | Deposit date: | 2015-01-27 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
2J7I
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETERODIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal Src Homology 3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J6O
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETEROTRIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
4AR6
 
 | | X-ray crystallographic structure of the reduced form perdeuterated Pyrococcus furiosus rubredoxin at 295 K (in quartz capillary) to 0.92 Angstroms resolution. | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Cuypers, M.G, Mason, S.A, Blakeley, M.P, Mitchell, E.P, Haertlein, M, Forsyth, V.T. | | Deposit date: | 2012-04-20 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Near-Atomic Resolution Neutron Crystallography on Perdeuterated Pyrococcus Furiosus Rubredoxin: Implication of Hydronium Ions and Protonation Equilibria and Hydronium Ions in Redox Changes
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
8AB0
 
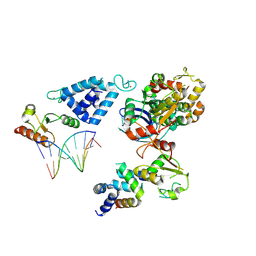 | | Complex of RecO-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA repair protein RecO, Oligo1, Oligo2, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2J3L
 
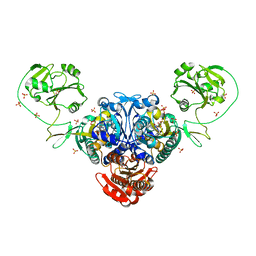 | | Prolyl-tRNA synthetase from Enterococcus faecalis complexed with a prolyl-adenylate analogue ('5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE) | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, PROLYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a Cis-Editing Domain.
Structure, 14, 2006
|
|
2QER
 
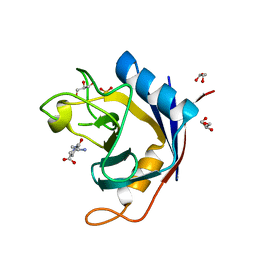 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 in the presence of dipeptide ala-pro | | Descriptor: | ALANINE, Cyclophilin-like protein, putative, ... | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 in the presence of dipeptide ala-pro.
To be Published
|
|
4AQF
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
2JEX
 
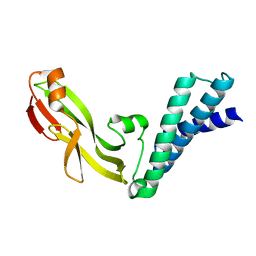 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-24 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
4AE5
 
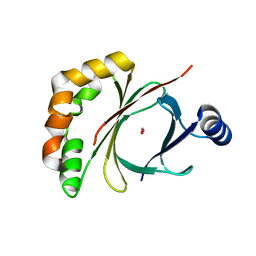 | |
2JC4
 
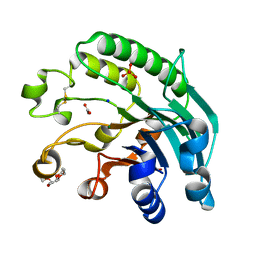 | | 3'-5' exonuclease (NExo) from Neisseria Meningitidis | | Descriptor: | ACETATE ION, DIHYDROGENPHOSPHATE ION, EXODEOXYRIBONUCLEASE III, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
2JC6
 
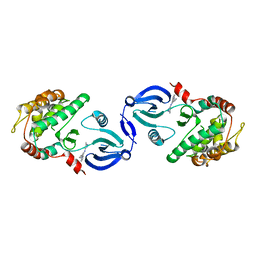 | | Crystal structure of human calmodulin-dependent protein kinase 1D | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE 1D, N-(5-METHYL-1H-PYRAZOL-3-YL)-2-PHENYLQUINAZOLIN-4-AMINE | | Authors: | Debreczeni, J.E, Rellos, P, Fedorov, O, Niesen, F.H, Bhatia, C, Shrestha, L, Salah, E, Smee, C, Colebrook, S, Berridge, G, Gileadi, O, Bunkoczi, G, Ugochukwu, E, Pike, A.C.W, von Delft, F, Knapp, S, Sundstrom, M, Weigelt, J, Arrowsmith, C.H, Edwards, A. | | Deposit date: | 2006-12-19 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase 1D
To be Published
|
|
4ADO
 
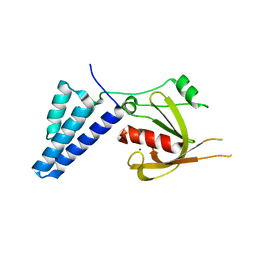 | | Fusidic acid resistance protein FusB | | Descriptor: | FAR1, ZINC ION | | Authors: | Guo, X, Peisker, K, Backbro, K, Chen, Y, Kiran, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2011-12-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Open Biol., 2, 2012
|
|
2JF7
 
 | | Structure of Strictosidine Glucosidase | | Descriptor: | STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
4B2P
 
 | | RadA C-terminal ATPase domain from Pyrococcus furiosus bound to GTP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
2QU8
 
 | | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Putative nucleolar GTP-binding protein 1 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Kozieradzki, I, Zhao, Y, Ravichandran, M, Shapiro, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Sukumar, D, Hassanali, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum.
To be Published
|
|
2JIL
 
 | | Crystal structure of 2nd PDZ domain of glutamate receptor interacting protein-1 (GRIP1) | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMATE RECEPTOR INTERACTING PROTEIN-1, THIOCYANATE ION | | Authors: | Tickle, J, Elkins, J, Pike, A.C.W, Cooper, C, Salah, E, Papagrigoriou, E, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of 2Nd Pdz Domain of Glutamate Receptor Interacting Protein-1 (Grip1)
To be Published
|
|
2JK2
 
 | | STRUCTURAL BASIS OF HUMAN TRIOSEPHOSPHATE ISOMERASE DEFICIENCY. CRYSTAL STRUCTURE OF THE WILD TYPE ENZYME. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, De Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | Deposit date: | 2008-06-22 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
2JEV
 
 | | Crystal structure of human spermine,spermidine acetyltransferase in complex with a bisubstrate analog (N1-acetylspermine-S-CoA). | | Descriptor: | (3R)-27-AMINO-3-HYDROXY-2,2-DIMETHYL-4,8,14-TRIOXO-12-THIA-5,9,15,19,24-PENTAAZAHEPTACOS-1-YL [(2S,3R,4S,5S)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, DIAMINE ACETYLTRANSFERASE 1 | | Authors: | Hegde, S.S, Chandler, J, Vetting, M.W, Yu, M, Blanchard, J.S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic and Structural Analysis of Human Spermidine/Spermine N(1)-Acetyltransferase.
Biochemistry, 46, 2007
|
|
4B0S
 
 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
4AY5
 
 | | Human O-GlcNAc transferase (OGT) in complex with UDP and glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GTAB1TIDE, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYL TRANSFERASE 110 KDA SUBUNIT, ... | | Authors: | Schimpl, M, Zheng, X, Blair, D.E, Schuettelkopf, A.W, Navratilova, I, Aristotelous, T, Ferenbach, A.T, Macnaughtan, M.A, Borodkin, V.S, van Aalten, D.M.F. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | O-Glcnac Transferase Invokes Nucleotide Sugar Pyrophosphate Participation in Catalysis
Nat.Chem.Biol., 8, 2012
|
|
4B87
 
 | | Crystal structure of human DNA cross-link repair 1A | | Descriptor: | 1,2-ETHANEDIOL, DNA CROSS-LINK REPAIR 1A PROTEIN, ZINC ION | | Authors: | Allerston, C.K, Berridge, G, Carpenter, E.P, Kochan, G, Krojer, T, Mahajan, P, Vollmar, M, Yue, W.W, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-08-24 | | Release date: | 2012-11-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of Human DNA Cross-Link Repair 1A
To be Published
|
|
2JIF
 
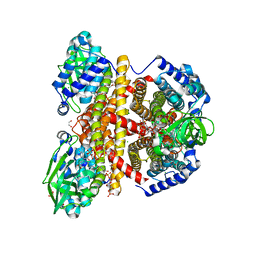 | | Structure of human short-branched chain acyl-CoA dehydrogenase (ACADSB) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A PERSULFIDE, ... | | Authors: | Pike, A.C.W, Hozjan, V, Smee, C, Niesen, F.H, Kavanagh, K.L, Umeano, C, Turnbull, A.P, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Oppermann, U. | | Deposit date: | 2007-02-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Short-Branched Chain Acyl-Coa Dehydrogenase
To be Published
|
|
4B2S
 
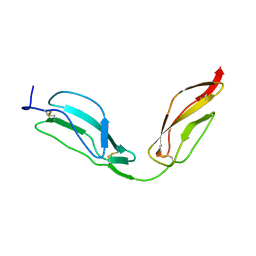 | | Solution structure of CCP modules 11-12 of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Makou, E, Mertens, H.D, Maciejewski, M, Soares, D.C, Matis, I, Schmidt, C.Q, Herbert, A.P, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ccp Modules 10-12 Illuminates Functional Architecture of the Complement Regulator, Factor H.
J.Mol.Biol., 424, 2012
|
|
