5OXT
 
 | |
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3OGR
 
 | | Complex structure of beta-galactosidase from Trichoderma reesei with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-17 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
6HYR
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT Q193C+MMTS | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3OH8
 
 | | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91 | | Descriptor: | Nucleoside-diphosphate sugar epimerase (SulA family) | | Authors: | Vorobiev, S, Lew, S, Kuzin, A, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structure of the nucleoside-diphosphate sugar epimerase from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR91.
To be Published
|
|
3OAU
 
 | | Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Non-Domain-Exchanged Forms, but only binds the HIV-1 Glycan Shield if Domain-Exchanged | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Huber, M, Wilson, I.A, Burton, D.R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged.
J.Virol., 84, 2010
|
|
6H9A
 
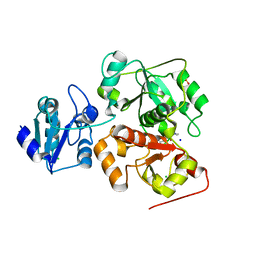 | | Crystal structure of anaerobic ergothioneine biosynthesis enzyme from Chlorobium limicola in complex with natural substrate trimethyl histidine. | | Descriptor: | CHLORIDE ION, N,N,N-trimethyl-histidine, SODIUM ION, ... | | Authors: | Leisinger, F, Burn, R, Meury, M, Lukat, P, Seebeck, F.P. | | Deposit date: | 2018-08-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Structural and Mechanistic Basis for Anaerobic Ergothioneine Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
3OGK
 
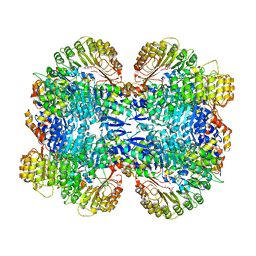 | | Structure of COI1-ASK1 in complex with coronatine and an incomplete JAZ1 degron | | Descriptor: | (1S,2S)-2-ethyl-1-({[(3aS,4S,6R,7aS)-6-ethyl-1-oxooctahydro-1H-inden-4-yl]carbonyl}amino)cyclopropanecarboxylic acid, Coronatine-insensitive protein 1, JAZ1 incomplete degron peptide, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
8BWU
 
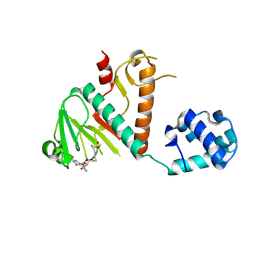 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | Deposit date: | 2022-12-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Application of established computational techniques to identify potential SARS-CoV-2 Nsp14-MTase inhibitors in low data regimes
Digit Discov, 2024
|
|
6HEH
 
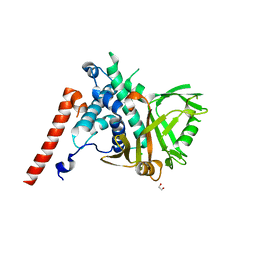 | | Structure of the catalytic domain of USP28 (insertion deleted) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28,Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HEI
 
 | |
6HEL
 
 | | Structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6HY9
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT Q193M | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-19 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6HYA
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT Q193L | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-19 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3OGX
 
 | | Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of heparin binding to camel peptidoglycan recognition protein-S
Int J Biochem Mol Biol, 3, 2012
|
|
6HYZ
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT K248C | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3OGS
 
 | | Complex structure of beta-galactosidase from Trichoderma reesei with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maksimainen, M, Rouvinen, J. | | Deposit date: | 2010-08-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Trichoderma reesei beta-galactosidase reveal conformational changes in the active site
J.Struct.Biol., 174, 2011
|
|
3OCI
 
 | | Crystal structure of TBP (TATA box binding protein) | | Descriptor: | 1,2-ETHANEDIOL, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Cui, S, Wollmann, P, Moldt, M, Hopfner, K.-P. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
6HTO
 
 | | Tryptophan lyase 'empty state' | | Descriptor: | 3-methyl-2-indolic acid synthase, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Amara, P, Mouesca, J.M, Bella, M, Martin, L, Saragaglia, C, Gambarelli, S, Nicolet, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Radical S-Adenosyl-l-methionine Tryptophan Lyase (NosL): How the Protein Controls the Carboxyl Radical •CO2-Migration.
J.Am.Chem.Soc., 140, 2018
|
|
6HU7
 
 | |
6HUF
 
 | | Coping with strong translational non-crystallographic symmetry and extreme anisotropy in molecular replacement with Phaser: human Rab27a | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-27A | | Authors: | Jamshidiha, M, Perez-Dorado, I, Murray, J.W, Tate, E.W, Cota, E, Read, R.J. | | Deposit date: | 2018-10-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Coping with strong translational noncrystallographic symmetry and extreme anisotropy in molecular replacement with Phaser: human Rab27a.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6I08
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT E243C-I201W | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-25 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HV1
 
 | |
6HYK
 
 | | NMR solution structure of the C/D box snoRNA U14 | | Descriptor: | RNA (31-MER) | | Authors: | Chagot, M.E, Quinternet, M, Rothe, B, Charpentier, B, Coutant, J, Manival, X, Lebars, I. | | Deposit date: | 2018-10-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The yeast C/D box snoRNA U14 adopts a "weak" K-turn like conformation recognized by the Snu13 core protein in solution.
Biochimie, 164, 2019
|
|
