3CC9
 
 | | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Putative farnesyl pyrophosphate synthase, SODIUM ION | | Authors: | Wernimont, A.K, Dunford, J, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Plasmodium vivax putative polyprenyl pyrophosphate synthase in complex with geranylgeranyl diphosphate.
To be Published
|
|
7W61
 
 | | Crystal structure of farnesol dehydrogenase from Helicoverpa armigera | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Farnesol dehydrogenase, ... | | Authors: | Kumar, R, Das, J, Mahto, J.K, Sharma, M, Kumar, P, Sharma, A.K. | | Deposit date: | 2021-12-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and molecular characterization of NADP + -farnesol dehydrogenase from cotton bollworm, Helicoverpaarmigera.
Insect Biochem.Mol.Biol., 147, 2022
|
|
3C8O
 
 | | The Crystal Structure of RraA from PAO1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Regulator of ribonuclease activity A, ... | | Authors: | Luo, M, Niu, S, Yin, Y, Huang, A, Wang, D. | | Deposit date: | 2008-02-12 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of RraA from PAO1
To be published
|
|
6IZB
 
 | | Flexible loop truncated human TCTP | | Descriptor: | Translationally-controlled tumor protein | | Authors: | Shin, D.H, Kim, M.S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Flexible loop truncated human TCTP
To Be Published
|
|
7W7S
 
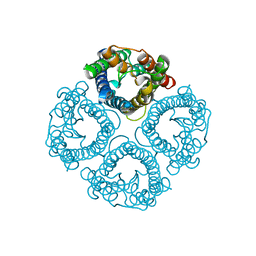 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
3S1T
 
 | |
3CAB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 7.0 | | Descriptor: | GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
7WBN
 
 | | PDB structure of RevCC | | Descriptor: | RevCC | | Authors: | Han, S, Kim, D, Kaur, M, Lim, Y.B, Barnwal, R.P. | | Deposit date: | 2021-12-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pseudo-Isolated alpha-Helix Platform for the Recognition of Deep and Narrow Targets.
J.Am.Chem.Soc., 144, 2022
|
|
3CAX
 
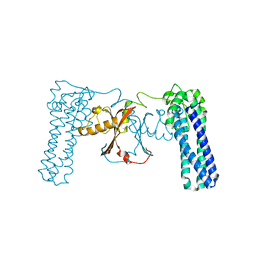 | | Crystal structure of uncharacterized protein PF0695 | | Descriptor: | Uncharacterized protein PF0695 | | Authors: | Ramagopal, U.A, Hu, S, Toro, R, Gilmore, M, Bain, K, Meyer, A.J, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of uncharacterized protein PF0695.
To be Published
|
|
7CSP
 
 | | Structure of Ephexin4 IDPSH | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WDR
 
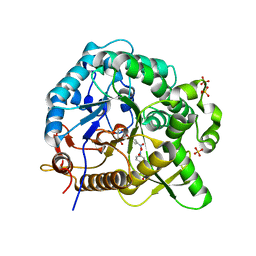 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, Beta-glucosidase, SULFATE ION | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7CSR
 
 | | Structure of Ephexin4 R676L | | Descriptor: | Rho guanine nucleotide exchange factor 16 | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WC2
 
 | | Cryo-EM structure of alphavirus, Getah virus | | Descriptor: | Spike glycoprotein E1, Spike glycoprotein E2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, M, Sun, Z.Z, Wang, J.F. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Implications for the pathogenicity and antigenicity of alpha viruses revealed by a 3.5 angstrom Cryo-EM structure of Getah virus
To Be Published
|
|
7CDM
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (4500 images from 2nd half of data set) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CSO
 
 | | Structure of Ephexin4 DH-PH-SH3 | | Descriptor: | Rho guanine nucleotide exchange factor 16, SULFATE ION | | Authors: | Zhang, M, Lin, L, Wang, C, Zhu, J. | | Deposit date: | 2020-08-15 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Double inhibition and activation mechanisms of Ephexin family RhoGEFs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WDP
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
3CDC
 
 | |
7CDN
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (9000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDP
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 42 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDQ
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 83 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7CDR
 
 | | Lysozyme room-temperature structure determined by SS-ROX combined with HAG method, 210 kGy (3000 images) | | Descriptor: | Lysozyme C, MALONATE ION, SODIUM ION | | Authors: | Hasegawa, K, Baba, S, Kawamura, T, Yamamoto, M, Kumasaka, T. | | Deposit date: | 2020-06-20 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of the data-collection strategy for room-temperature micro-crystallography studied by serial synchrotron rotation crystallography combined with the humid air and glue-coating method.
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7WDS
 
 | | Crystal structures of MeBglD2 in complex with various saccharides | | Descriptor: | Beta-glucosidase, SULFATE ION, beta-D-xylopyranose | | Authors: | Watanabe, M, Matsuzawa, T, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-12-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of metagenomic beta-glycosidase MeBglD2 in complex with various saccharides
Appl.Microbiol.Biotechnol., 106, 2022
|
|
7C3G
 
 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with a bicyclic pyrazole inhibitor RK-73134 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-1, SULFATE ION, ... | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Novel bicyclic pyrazoles as potent ALK2 (R206H) inhibitors for the treatment of fibrodysplasia ossificans progressiva.
Bioorg.Med.Chem.Lett., 38, 2021
|
|
3CDU
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
3C9N
 
 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
