3AYN
 
 | | Crystal structure of squid isorhodopsin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PALMITIC ACID, RETINAL, ... | | Authors: | Murakami, M, Kouyama, T. | | Deposit date: | 2011-05-09 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic Analysis of the Primary Photochemical Reaction of Squid Rhodopsin
J.Mol.Biol., 413, 2011
|
|
9J2N
 
 | | Cryo-EM structure of the human glucose transporter, GLUT7 in outward-facing open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 2, facilitated glucose transporter member 7 | | Authors: | Lee, S.S, Kim, S, Jin, M.S. | | Deposit date: | 2024-08-07 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human glucose transporter GLUT7.
Biochem.Biophys.Res.Commun., 738, 2024
|
|
3AWO
 
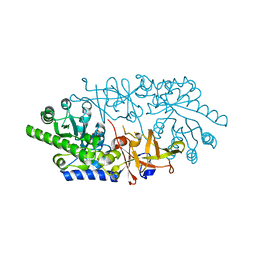 | | Crystal structure of D-serine dehydratase in complex with D-serine from chicken kidney (EDTA-treated) | | Descriptor: | D-SERINE, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3AWR
 
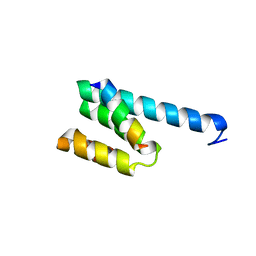 | |
3AYU
 
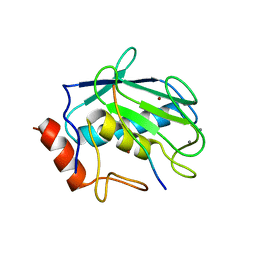 | | Crystal structure of MMP-2 active site mutant in complex with APP-drived decapeptide inhibitor | | Descriptor: | 72 kDa type IV collagenase, Amyloid beta A4 protein, CALCIUM ION, ... | | Authors: | Hashimoto, H, Takeuchi, T, Komatsu, K, Miyazaki, K, Sato, M, Higashi, S. | | Deposit date: | 2011-05-17 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for matrix metalloproteinase-2 (MMP-2)-selective inhibitory action of {beta}-amyloid precursor protein-derived inhibitor
J.Biol.Chem., 2011
|
|
7BJT
 
 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | Alginate lyase, family PL17, CALCIUM ION, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
7BM6
 
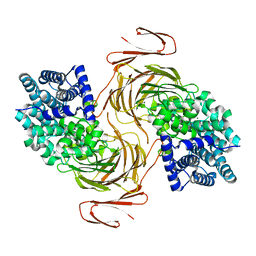 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid, Alginate lyase, family PL17, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
3AWI
 
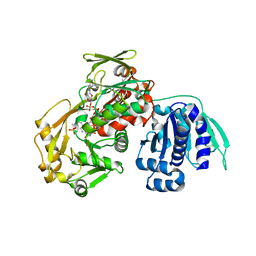 | | Bifunctional tRNA modification enzyme MnmC from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein mnmC | | Authors: | Kitamura, A, Sengoku, T, Nishimoto, M, Yokoyama, S, Bessho, Y. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the bifunctional tRNA modification enzyme MnmC from Escherichia coli
Protein Sci., 2011
|
|
7FLT
 
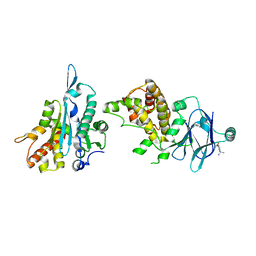 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G04 from the F2X-Universal Library | | Descriptor: | (1S)-1-(3-fluoro-4-methoxyphenyl)ethan-1-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7V56
 
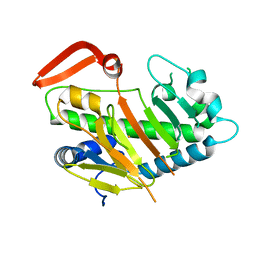 | | Structure of AdaV | | Descriptor: | AdaV, FE (III) ION | | Authors: | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7FKC
 
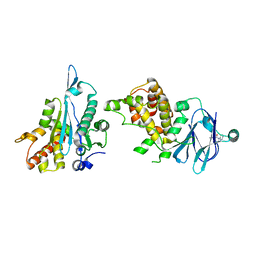 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04C10 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N,N~2~-dimethyl-N-(pyridin-2-yl)glycinamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7V9Z
 
 | |
7FNT
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07E12 from the F2X-Universal Library | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}ethan-1-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FNQ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07E01 from the F2X-Universal Library | | Descriptor: | (3S)-3-methyl-5-oxo-5-[(1,3-thiazol-2-yl)amino]pentanoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLP
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05F11 from the F2X-Universal Library | | Descriptor: | 2-methoxybenzamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FN6
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P06H06 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ethyl (3-methylanilino)(oxo)acetate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOR
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08C12 from the F2X-Universal Library | | Descriptor: | (2S)-1-(2-bromophenoxy)-3-(pyrrolidin-1-yl)propan-2-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7VA3
 
 | |
7FOU
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08E01 from the F2X-Universal Library | | Descriptor: | (1R)-1-(4-fluorophenyl)-2-[(propan-2-yl)amino]ethan-1-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FP3
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08F11 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N,N~2~-diethyl-N~2~-(2-methylpyrimidin-4-yl)glycinamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FK3
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A09 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, methyl N-(benzenesulfonyl)-N-methylglycinate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FL7
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05A01 from the F2X-Universal Library | | Descriptor: | (2R)-(4-chlorophenyl)(morpholin-4-yl)acetic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLX
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G08 from the F2X-Universal Library | | Descriptor: | 2-chlorobenzohydrazide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FP0
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08F02 from the F2X-Universal Library | | Descriptor: | 1-cyclopropylimidazolidin-2-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
