8BOG
 
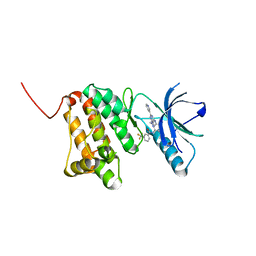 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 7 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-[4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOM
 
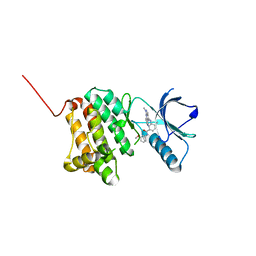 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 14 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3-methoxyphenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOD
 
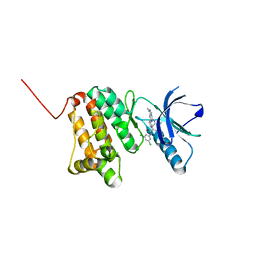 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 20 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-[3,5-bis(chloranyl)phenyl]-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOI
 
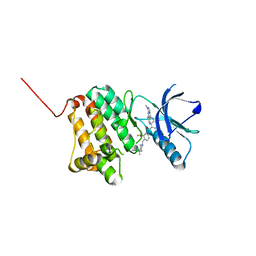 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 9 | | Descriptor: | 4-methyl-~{N}-[4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide, Ephrin type-A receptor 2 | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOK
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 11 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3,5-dimethoxyphenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOH
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 8 | | Descriptor: | 4-methyl-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide, Ephrin type-A receptor 2 | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOF
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 12 | | Descriptor: | 4-methyl-~{N}-(3-methylphenyl)-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide, Ephrin type-A receptor 2 | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
4BBO
 
 | | Crystal structure of core-bradavidin | | Descriptor: | ACETATE ION, BIOTIN, BLR5658 PROTEIN, ... | | Authors: | Airenne, T.T, Johnson, M.S, Maatta, J.A.E, Hytonen, V.H, Kulomaa, M.S. | | Deposit date: | 2012-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Core-Bradavidin
To be Published
|
|
4K89
 
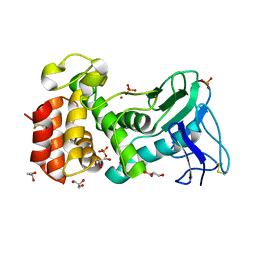 | | Crystal structure of Pseudomonas aeruginosa strain K solvent tolerant elastase | | Descriptor: | CALCIUM ION, GLYCEROL, Organic solvent tolerant elastase, ... | | Authors: | Ali, M.S.M, Said, Z.S.A.M, Rahman, R.N.Z.R.A, Basri, M, Salleh, A.B. | | Deposit date: | 2013-04-18 | | Release date: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure analysia of solvent tolerant elastase strain K
To be Published
|
|
9BEZ
 
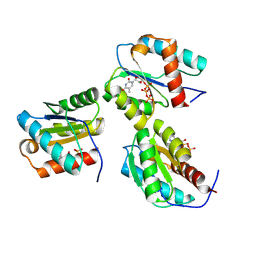 | | MID domain of human Argo2 bound to RNA | | Descriptor: | Protein argonaute-2, [(3~{S},4~{R},5~{R})-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-4-oxidanyl-oxolan-3-yl] [oxidanyl(phosphonooxy)phosphoryl] hydrogen phosphate | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9EZY
 
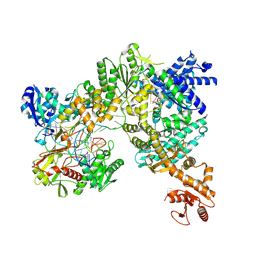 | | Vibrio cholerae DdmD-DdmE holo complex | | Descriptor: | 14 nucleotide DNA guide with terminal 5' phosphate, Helicase/UvrB N-terminal domain-containing protein, MAGNESIUM ION, ... | | Authors: | Loeff, L, Jinek, M. | | Deposit date: | 2024-04-14 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular mechanism of plasmid elimination by the DdmDE defense system.
Science, 385, 2024
|
|
9BI8
 
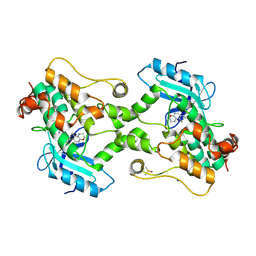 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (3R,4S)-4-methyloxolan-3-yl [(6P)-8-amino-7-fluoro-6-(8-methyl-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)isoquinolin-3-yl]carbamate, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase kinase 1, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-22 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
6L9F
 
 | |
6L5W
 
 | | Carbonmonoxy human hemoglobin C in the R quaternary structure at 140 K: Light (2 min) | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4R15
 
 | | High-resolution crystal structure of Z-DNA in complex with Cr3+ cations | | Descriptor: | CHROMIUM ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2014-08-04 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations.
J.Biol.Inorg.Chem., 20, 2015
|
|
6LB0
 
 | |
9B9R
 
 | |
9CE4
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 indazole inhibitor compound 6 | | Descriptor: | (1S,4r)-4-[(1P)-5-chloro-1-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-6-yl]-1-(oxetan-3-yl)piperidin-1-ium, 1,2-ETHANEDIOL, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L, Zebisch, M, Henry, C, Barker, J.J. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery and Optimization of N-Heteroaryl Indazole LRRK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
6LJG
 
 | | Crassostrea gigas ferritin mutant-D119G | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Li, H, Zang, J, Tan, X, Wang, Z, Du, M. | | Deposit date: | 2019-12-15 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crassostrea gigas ferritin mutant-D119G
To Be Published
|
|
6LBU
 
 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
9BJ1
 
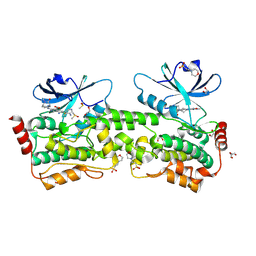 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
5OD3
 
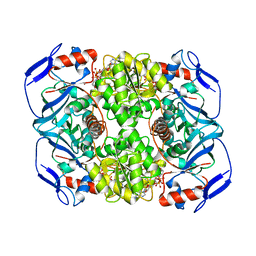 | | Crystal structure of R. ruber ADH-A, mutant Y54G, L119Y | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereo- and Regioselectivity in Catalyzed Transformation of a 1,2-Disubstituted Vicinal Diol and the Corresponding Diketone by Wild Type and Laboratory Evolved Alcohol Dehydrogenases
Acs Catalysis, 8, 2018
|
|
9BS3
 
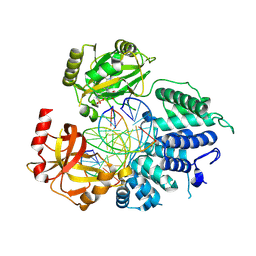 | | Wild type DNA Ligase 1 with 5'-rG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | KanalElamparithi, B, Caglayan, M. | | Deposit date: | 2024-05-12 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of LIG1 uncover the mechanism of sugar discrimination against 5'-RNA-DNA junctions during ribonucleotide excision repair.
J.Biol.Chem., 300, 2024
|
|
4UTN
 
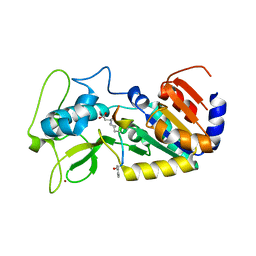 | | Crystal structure of zebrafish Sirtuin 5 in complex with succinylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-21 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
9BS4
 
 | | DNA Ligase 1 E346A/E592A double mutant with 5'-rG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | KanalElamparithi, B, Caglayan, M. | | Deposit date: | 2024-05-12 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of LIG1 uncover the mechanism of sugar discrimination against 5'-RNA-DNA junctions during ribonucleotide excision repair.
J.Biol.Chem., 300, 2024
|
|
