1T93
 
 | | Evidence for Multiple Substrate Recognition and Molecular Mechanism of C-C reaction by Cytochrome P450 CYP158A2 from Streptomyces Coelicolor A3(2) | | 分子名称: | FLAVIOLIN, PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | 著者 | Zhao, B, Sundaramoorthy, M, Waterman, M.R. | | 登録日 | 2004-05-14 | | 公開日 | 2005-01-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2.
J.Biol.Chem., 280, 2005
|
|
6RGO
 
 | |
7PCX
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | 分子名称: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | 著者 | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | 登録日 | 2021-08-04 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
112L
 
 | |
110L
 
 | |
6RXQ
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after soaking | | 分子名称: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
111L
 
 | |
113L
 
 | |
4ZO6
 
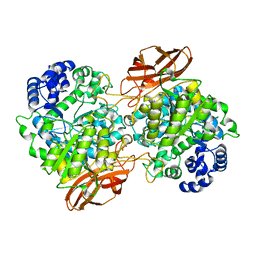 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with cellobiose | | 分子名称: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
8F3K
 
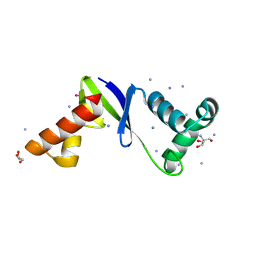 | | Anti-CRISPR protein AcrIIC5 inhibits CRISPR-Cas9 by acting as a DNA mimic | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACRIIC5Nch, AMMONIUM ION, ... | | 著者 | Shah, M, Sungwon, H, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-04-12 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Anti-CRISPR Protein AcrIIC5 Inhibits CRISPR-Cas9 by Occupying the Target DNA Binding Pocket.
J.Mol.Biol., 435, 2023
|
|
115L
 
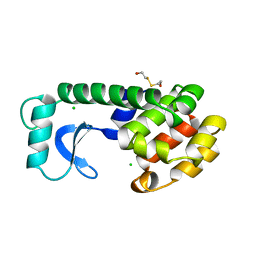 | |
114L
 
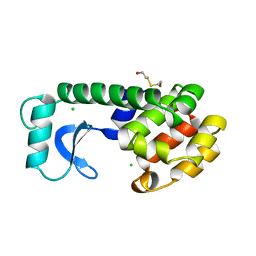 | |
3LK2
 
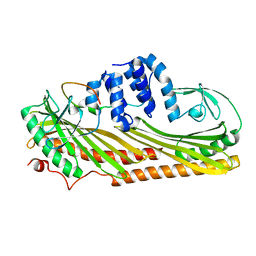 | | Crystal structure of CapZ bound to the uncapping motif from CARMIL | | 分子名称: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | 著者 | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Aguda, A.H, Larsson, M, Cooper, J.A, Robinson, R.C. | | 登録日 | 2010-01-27 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
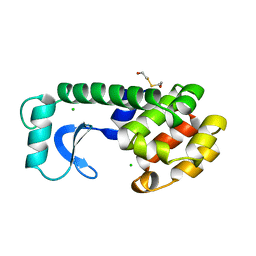
119L
 
 | |
6RLE
 
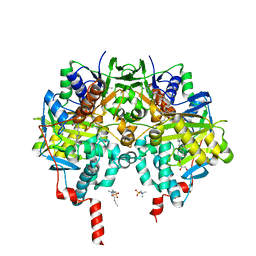 | | Crystal structure of human monoamine oxidase B in complex with styrylpiperidine analogue 97 | | 分子名称: | 4-[2-(4-propan-2-ylphenyl)ethyl]-1-[(~{E})-prop-1-enyl]piperidine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Iacovino, L.G, Knez, D, Colettis, N, Sova, M, Pislar, A, Higgs, J, Kamecki, F, Mangialavori, I, Dolsak, A, Zakelj, S, Trontelj, J, Kos, J, Marder, N.M, Gobec, S, Binda, C. | | 登録日 | 2019-05-02 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B.
J.Med.Chem., 63, 2020
|
|
1SS6
 
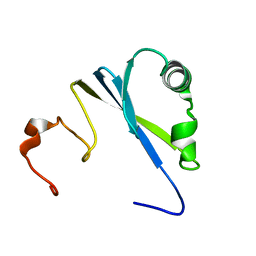 | | Solution structure of SEP domain from human p47 | | 分子名称: | NSFL1 cofactor p47 | | 著者 | Soukenik, M, Leidert, M, Sievert, V, Buessow, K, Leitner, D, Labudde, D, Ball, L.J, Oschkinat, H. | | 登録日 | 2004-03-23 | | 公開日 | 2004-11-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L
FEBS Lett., 576, 2004
|
|
118L
 
 | |
120L
 
 | |
3LLH
 
 | | Crystal structure of the first dsRBD of TAR RNA-binding protein 2 | | 分子名称: | MALONATE ION, RISC-loading complex subunit TARBP2 | | 著者 | Yamashita, S, Kawazoe, M, Takemoto, C, Sekine, S, Wakiyama, M, Yokoyama, S. | | 登録日 | 2010-01-29 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | The structures of dsRBDs of human TRBP
To be Published
|
|
8F23
 
 | | The crystal structure of a rationally designed zinc sensor based on maltose binding protein - Apo conformation | | 分子名称: | Zinc Sensor protein | | 著者 | Zhao, Z, Zhou, M, Zemerov, S.d, Marmorstein, R, Dmochowski, I.J. | | 登録日 | 2022-11-06 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Rational design of a genetically encoded NMR zinc sensor.
Chem Sci, 14, 2023
|
|
4ZO7
 
 | | Crystal structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with gentiobiose | | 分子名称: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
122L
 
 | |
4KMI
 
 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with PO4 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION | | 著者 | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | 登録日 | 2013-05-08 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
8EZD
 
 | |
123L
 
 | |
