7CIM
 
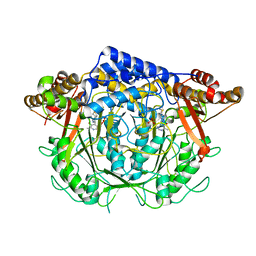 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (geminal diamine form). | | 分子名称: | L-methionine decarboxylase, [6-methyl-4-[(3-methylsulfanylpropylamino)methyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
8OPI
 
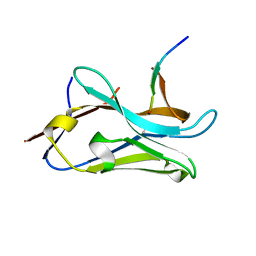 | | VHH Z70 mutant 1 in interaction with PHF6 Tau peptide | | 分子名称: | PHF6 Tau peptide, VHH Z70 mutant 1 | | 著者 | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | 登録日 | 2023-04-07 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
7CTR
 
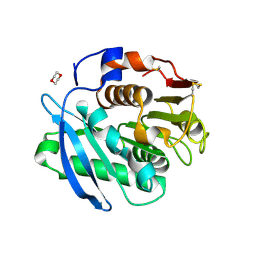 | | Closed form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein | | 著者 | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | 登録日 | 2020-08-20 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
7C3N
 
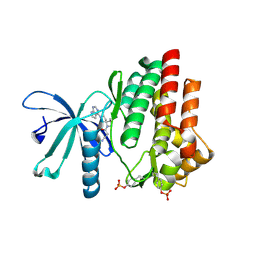 | | Crystal structure of JAK3 in complex with Delgocitinib | | 分子名称: | 3-[(3S,4R)-3-methyl-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,7-diazaspiro[3.4]octan-1-yl]-3-oxidanylidene-propanenitrile, Tyrosine-protein kinase JAK3 | | 著者 | Doi, S, Otira, T, Kikuwaka, M, Nomura, A, Noji, S, Adachi, T. | | 登録日 | 2020-05-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders.
J.Med.Chem., 63, 2020
|
|
8OP0
 
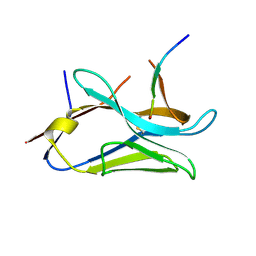 | | VHH Z70 mutant 20 in interaction with PHF6 Tau peptide | | 分子名称: | Tau PHF6 peptide, VHH Z70 mutant 20 | | 著者 | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | 登録日 | 2023-04-06 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
7CBO
 
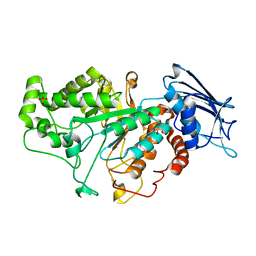 | | Crystal structure of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila in complex with GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | 著者 | Xu, W, Wang, M, Zhang, M. | | 登録日 | 2020-06-13 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and biochemical analyses of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila involved in mucin degradation.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
5LOU
 
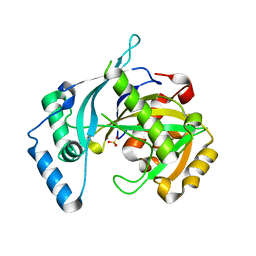 | | human NUDT22 | | 分子名称: | Nucleoside diphosphate-linked moiety X motif 22, PHOSPHATE ION | | 著者 | Carter, M, Stenmark, P. | | 登録日 | 2016-08-10 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Human NUDT22 Is a UDP-Glucose/Galactose Hydrolase Exhibiting a Unique Structural Fold.
Structure, 26, 2018
|
|
1GBR
 
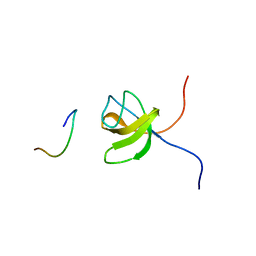 | | ORIENTATION OF PEPTIDE FRAGMENTS FROM SOS PROTEINS BOUND TO THE N-TERMINAL SH3 DOMAIN OF GRB2 DETERMINED BY NMR SPECTROSCOPY | | 分子名称: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, SOS-A PEPTIDE | | 著者 | Wittekind, M, Mapelli, C, Farmer, B.T, Suen, K.-L, Goldfarb, V, Tsao, J, Lavoie, T, Barbacid, M, Meyers, C.A, Mueller, L. | | 登録日 | 1994-08-12 | | 公開日 | 1995-01-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy.
Biochemistry, 33, 1994
|
|
7C6A
 
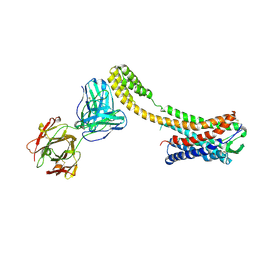 | | Crystal structure of AT2R-BRIL and SRP2070_Fab complex | | 分子名称: | IgG Light Chain, IgG heavy chain, SAR1, ... | | 著者 | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | 登録日 | 2020-05-21 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
3N5K
 
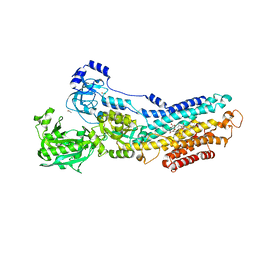 | | Structure Of The (Sr)Ca2+-ATPase E2-AlF4- Form | | 分子名称: | ACETATE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | 著者 | Bublitz, M, Olesen, C, Poulsen, H, Morth, J.P, Moller, J.V, Nissen, P. | | 登録日 | 2010-05-25 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Ion pathways in the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 288, 2013
|
|
6H6T
 
 | |
5G2H
 
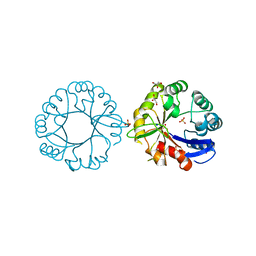 | | S. enterica HisA with mutation L169R | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SODIUM ION, ... | | 著者 | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | 登録日 | 2016-04-08 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3N99
 
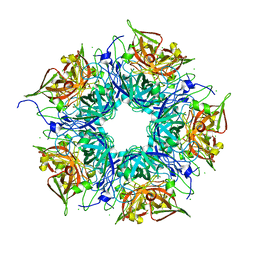 | | Crystal structure of TM1086 | | 分子名称: | CHLORIDE ION, uncharacterized protein TM1086 | | 著者 | Chruszcz, M, Domagalski, M.J, Wang, S, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-05-28 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystal structure of TM1086
To be Published
|
|
3NDS
 
 | | Crystal structure of engineered Naja Nigricollis toxin alpha | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, SULFATE ION, Short neurotoxin 1 | | 著者 | Stura, E.A, Drevet, P, Gregory, G, Gallopin, M, Vandame, M. | | 登録日 | 2010-06-08 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Conformational exchange is critical for the productivity of an oxidative folding intermediate with buried free cysteines.
J.Mol.Biol., 403, 2010
|
|
3NHP
 
 | |
5LWM
 
 | | Crystal structure of JAK3 in complex with Compound 4 (FM381) | | 分子名称: | 1,2-ETHANEDIOL, 1-phenylurea, 2-cyano-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl)furan-2-yl]-~{N},~{N}-dimethyl-prop-2-enamide, ... | | 著者 | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-18 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
3NGO
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | 分子名称: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | 著者 | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | 登録日 | 2010-06-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
7CBN
 
 | |
6HBN
 
 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA/CSKN2A1 GENE PRODUCT) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | 分子名称: | 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha, ... | | 著者 | Niefind, K, Hochscherf, J, Dimper, V, Witulski, B, Lindenblatt, D, Jose, J, Le Borgne, M. | | 登録日 | 2018-08-10 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
5WOP
 
 | | High Resolution Structure of Mutant CA09-PB2cap | | 分子名称: | GLYCEROL, Polymerase PB2 | | 著者 | Constantinides, A.E, Gumpper, R.H, Severin, C, Luo, M. | | 登録日 | 2017-08-02 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | High-resolution structure of the Influenza A virus PB2cap binding domain illuminates the changes induced by ligand binding.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1GAB
 
 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | PROTEIN PAB | | 著者 | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | 登録日 | 1996-12-30 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
5GN6
 
 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17b | | 分子名称: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | 著者 | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | 登録日 | 2016-07-19 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
5G1Y
 
 | | S. enterica HisA mutant D10G, dup13-15,V14:2M, Q24L, G102 | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SULFATE ION | | 著者 | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | 登録日 | 2016-04-01 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3M5Q
 
 | | 0.93 A Structure of Manganese-Bound Manganese Peroxidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | 著者 | Sundaramoorthy, M, Gold, M.H, Poulos, T.L. | | 登録日 | 2010-03-12 | | 公開日 | 2010-04-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | Ultrahigh (0.93A) resolution structure of manganese peroxidase from Phanerochaete chrysosporium: implications for the catalytic mechanism.
J.Inorg.Biochem., 104, 2010
|
|
5N18
 
 | | Second Bromodomain (BD2) from Candida albicans Bdf1 bound to an imidazopyridine (compound 2) | | 分子名称: | 4-[8-methyl-3-[(4-methylphenyl)amino]imidazo[1,2-a]pyridin-2-yl]phenol, Bromodomain-containing factor 1, GLYCEROL | | 著者 | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | 登録日 | 2017-02-05 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
