7GRO
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-11 | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6K55
 
 | | Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide | | 分子名称: | Glucanase, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | 登録日 | 2019-05-28 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.883 Å) | | 主引用文献 | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
7GRM
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-9 | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRQ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-13 | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRV
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-18 | | 分子名称: | (2S)-2-(2-fluorophenyl)-1,3-thiazolidin-4-one, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GS1
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-24 | | 分子名称: | 2-cyano-~{N}-cyclohexyl-ethanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-10 | | 分子名称: | 2-[(3S)-pyrrolidin-3-yl]-5-(trifluoromethyl)-1H-benzimidazole, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6K6J
 
 | | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion | | 分子名称: | BROMIDE ION, Cyanobacterial chloride importer, OLEIC ACID, ... | | 著者 | Yun, J.H, Park, J.H, Jin, Z, Ohki, M, Wang, Y, Lupala, C.S, Kim, M, Liu, H, Park, S.Y, Lee, W. | | 登録日 | 2019-06-03 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens with Bromide ion
To Be Published
|
|
7GS0
 
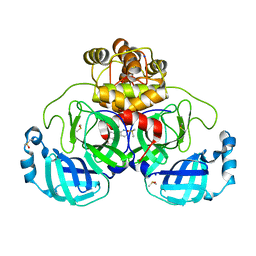 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-23 | | 分子名称: | (pyridin-2-yl)(quinolin-2-yl)methanone, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRS
 
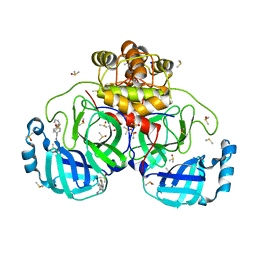 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-15 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, SODIUM ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2CXL
 
 | | RUN domain of Rap2 interacting protein x, crystallized in I422 space group | | 分子名称: | rap2 interacting protein x | | 著者 | Kukimoto-Niino, M, Umehara, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-06-30 | | 公開日 | 2005-12-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
7GRF
 
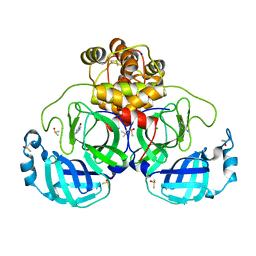 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-2 | | 分子名称: | 3C-like proteinase nsp5, 5-bromopyridin-3-amine, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRJ
 
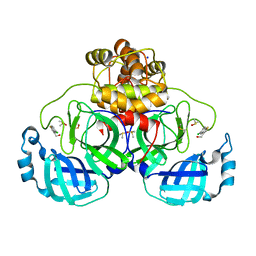 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-6 | | 分子名称: | (5-chloro-1-benzothiophen-3-yl)methanol, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6UFJ
 
 | | Pistol ribozyme product crystal structure | | 分子名称: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | 著者 | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | 登録日 | 2019-09-24 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.645 Å) | | 主引用文献 | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7GRW
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-19 | | 分子名称: | (2S)-N-(3,5-dichlorophenyl)-2-hydroxypropanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-20 | | 分子名称: | 1-(2,4-difluorophenyl)pyrrolidine-2,5-dione, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7GRU
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with cpd-17 | | 分子名称: | 3-(4-chlorophenyl)-1-methyl-1H-pyrazol-5-amine, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Huang, C.-Y, Metz, A, Sharpe, M, Sweeney, A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Fragment-based screening targeting an open form of the SARS-CoV-2 main protease binding pocket.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
2Q6G
 
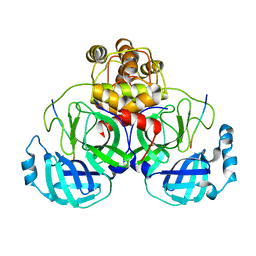 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | 分子名称: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2CZL
 
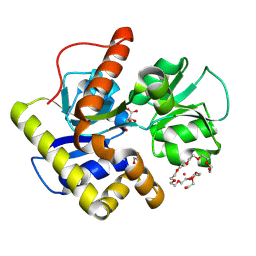 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 (Cys11 modified with beta-mercaptoethanol) | | 分子名称: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | 著者 | Arai, R, Nishino, A, Nagano, K, Kamo-Uchikubo, T, Nishimoto, M, Toyama, M, Terada, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-13 | | 公開日 | 2006-01-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
2D00
 
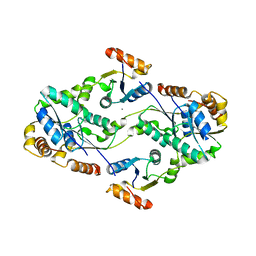 | | Subunit F of V-type ATPase/synthase | | 分子名称: | CALCIUM ION, V-type ATP synthase subunit F | | 著者 | Makyio, H, Iino, R, Ikeda, C, Imamura, H, Tamakoshi, M, Iwata, M, Stock, D, Bernal, R.A, Carpenter, E.P, Yoshida, M, Yokoyama, K, Iwata, S. | | 登録日 | 2005-07-21 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus
Embo J., 24, 2005
|
|
3NHK
 
 | |
5H17
 
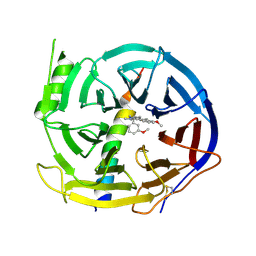 | | EED in complex with PRC2 allosteric inhibitor EED210 | | 分子名称: | (3R,4aS,10aS)-6-methoxy-3-[(3-methoxyphenyl)methyl]-1-methyl-3,4,4a,5,10,10a-hexahydro-2H-benzo[g]quinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | 著者 | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | 登録日 | 2016-10-08 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
6UEY
 
 | | Pistol ribozyme transition-state analog vanadate | | 分子名称: | MAGNESIUM ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | 著者 | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | 登録日 | 2019-09-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2CWI
 
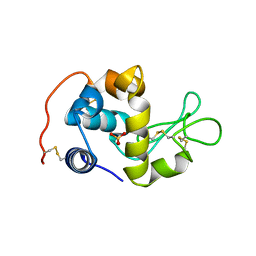 | | X-ray crystal structure analysis of recombinant wild-type canine milk lysozyme (apo-type) | | 分子名称: | Lysozyme C, milk isozyme, SULFATE ION | | 著者 | Akieda, D, Yasui, M, Aizawa, T, Yao, M, Watanabe, N, Tanaka, I, Demura, M, Kawano, K, Nitta, K. | | 登録日 | 2005-06-20 | | 公開日 | 2006-06-20 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | Construction of an expression system of canine milk lysozyme in the methylotrophic yeast Pichia pastoris
To be Published
|
|
2CYB
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Archaeoglobus fulgidus | | 分子名称: | TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-06 | | 公開日 | 2005-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
