5S35
 
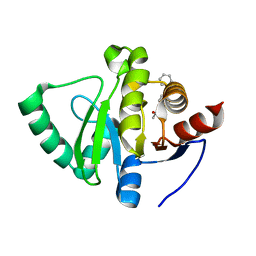 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z68404778 | | Descriptor: | Non-structural protein 3, ~{N}-(4-phenylazanylphenyl)ethanamide | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3L
 
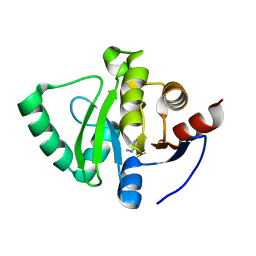 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z54628578 | | Descriptor: | N-methylpyrimidin-2-amine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.091 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S42
 
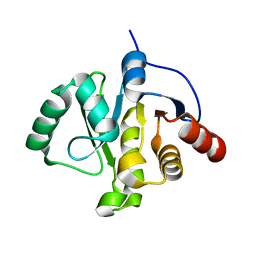 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with NCL-00023833 | | Descriptor: | 4-bromanyl-1,8-naphthyridine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S1A
 
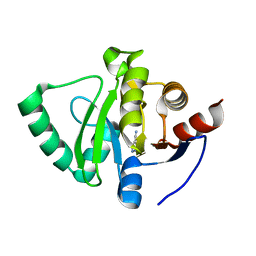 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-43406 | | Descriptor: | 5-amino-3-methyl-1H-pyrazole-4-carbonitrile, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S6X
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2889976755 | | Descriptor: | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S2D
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z369936976 | | Descriptor: | N'-cyclopropyl-N-methyl-N-[(5-methyl-1,2-oxazol-3-yl)methyl]urea, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S22
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z145120524 | | Descriptor: | 2H-1-benzopyran-3-carboxamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.175 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2T
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z26781964 | | Descriptor: | N-[(1H-benzimidazol-2-yl)methyl]furan-2-carboxamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.108 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2J
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z509756472 | | Descriptor: | N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.111 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z126932614 | | Descriptor: | 2-[(methylsulfonyl)methyl]-1H-benzimidazole, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S38
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1745658474 | | Descriptor: | 2-(trifluoromethyl)pyrimidine-5-carboxamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.072 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3F
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z57446103 | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-propyl-2H-tetrazol-5-yl)furan-2-carboxamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3J
 
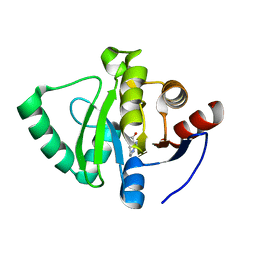 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1324853681 | | Descriptor: | (8S)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-8-carboxamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.087 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3V
 
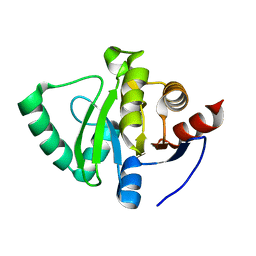 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0120 | | Descriptor: | (2R)-1',4'-dihydro-2'H-spiro[pyrrolidine-2,3'-quinolin]-2'-one, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S40
 
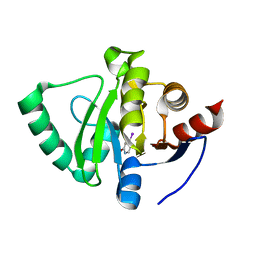 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with NCL-00023824 | | Descriptor: | 4-iodanyl-3~{H}-pyridin-2-one, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.187 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4B
 
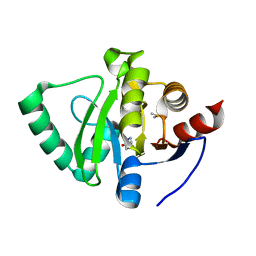 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z3219959731 | | Descriptor: | DIMETHYL SULFOXIDE, Non-structural protein 3, pyridazin-3(2H)-one | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.185 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4G
 
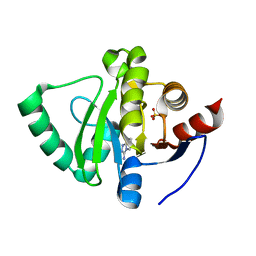 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | Descriptor: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S70
 
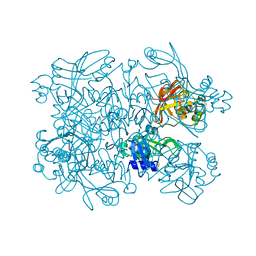 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | Descriptor: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
1VDP
 
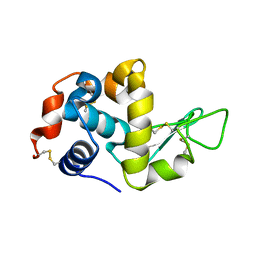 | | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space
to be published
|
|
1VPR
 
 | |
1VTF
 
 | |
1VND
 
 | | VND/NK-2 PROTEIN (HOMEODOMAIN), NMR | | Descriptor: | VND/NK-2 PROTEIN | | Authors: | Tsao, D.H.H, Gruschus, J.M, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1996-05-22 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the NK-2 homeodomain from Drosophila.
J.Mol.Biol., 251, 1995
|
|
2AOT
 
 | | Histamine Methyltransferase Complexed with the Antihistamine Drug Diphenhydramine | | Descriptor: | Histamine N-methyltransferase, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
1VM1
 
 | | STRUCTURE OF SHV-1 BETA-LACTAMASE INHIBITED BY TAZOBACTAM | | Descriptor: | ACRYLIC ACID, BETA-LACTAMASE SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, ... | | Authors: | Kuzin, A.P, Nukaga, M, Nukaga, Y, Hujer, A, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-08-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Inhibition of the SHV-1 beta-lactamase by sulfones: crystallographic observation of two reaction intermediates with tazobactam.
Biochemistry, 40, 2001
|
|
3TRP
 
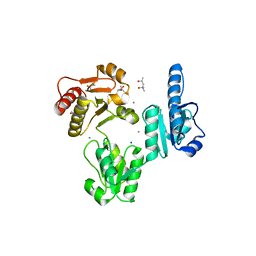 | | Crystal structure of recombinant rabbit skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Munske, G.R, Nissen, M.S, Kang, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8817 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
