1OJW
 
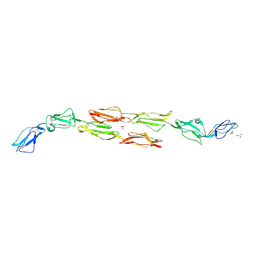 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, SULFATE ION | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, C.M, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7OG0
 
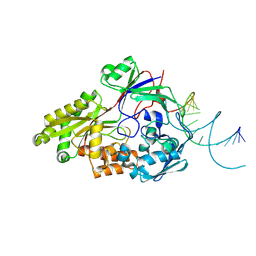 | | Nontypeable Haemophillus influenzae SapA in open and closed conformations, in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFZ
 
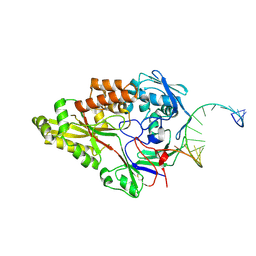 | | Nontypeable Haemophillus influenzae SapA in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFW
 
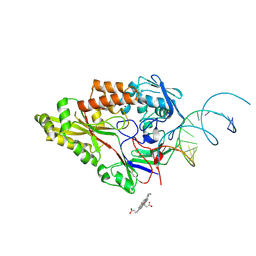 | | Nontypeable Haemophillus influenzae SapA in complex with heme | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
1OJY
 
 | | Decay accelerating factor (cd55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK2
 
 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OJV
 
 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, C.M, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK1
 
 | | Decay accelerating factor (cd55) : the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK3
 
 | | Decay accelerating factor (cd55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK9
 
 | | Decay accelerating factor (CD55): The structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, CHLORIDE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-21 | | Release date: | 2004-01-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1YDE
 
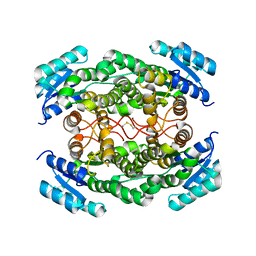 | | Crystal Structure of Human Retinal Short-Chain Dehydrogenase/Reductase 3 | | Descriptor: | Retinal dehydrogenase/reductase 3 | | Authors: | Lukacik, P, Bunkozci, G, Kavanagh, K, Sundstrom, M, Arrowsmith, C, Edwards, A, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-18 | | Last modified: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of human orphan DHRS10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity.
Biochem.J., 402, 2007
|
|
1YB1
 
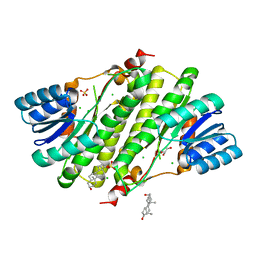 | | Crystal structure of human 17-beta-hydroxysteroid dehydrogenase type XI | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase type XI, Androsterone, CHLORIDE ION, ... | | Authors: | Lukacik, P, Bunkoczi, G, Kavanagh, K, Ng, S, von Delft, F, Bray, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human 17-beta-hydroxysteroid dehydrogenase type XI.
To be Published
|
|
4EPA
 
 | |
4EPF
 
 | |
1ZSY
 
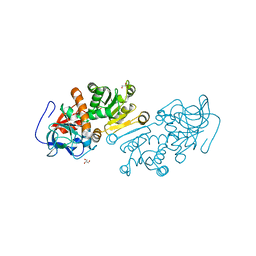 | | The structure of human mitochondrial 2-enoyl thioester reductase (CGI-63) | | Descriptor: | GLYCEROL, MITOCHONDRIAL 2-ENOYL THIOESTER REDUCTASE, SULFATE ION | | Authors: | Lukacik, P, Shafqat, N, Kavanagh, K.L, Johansson, C, Smee, C, Edwards, A, Arrowsmith, C, Sundstrom, M, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of human mitochondrial 2-enoyl thioester reductase (CGI-63)
To be Published
|
|
1ZBQ
 
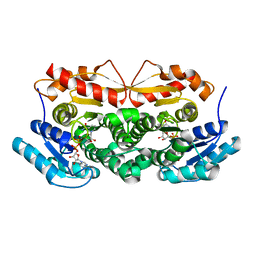 | | Crystal Structure Of Human 17-Beta-Hydroxysteroid Dehydrogenase Type 4 In Complex With NAD | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 4, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lukacik, P, Shafqat, N, Kavanagh, K, Bray, J, von Delft, F, Edwards, A, Arrowsmith, C, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure Of Human 17-Beta-Hydroxysteroid Dehydrogenase Type 4 In Complex With NAD
To be Published
|
|
4YZ4
 
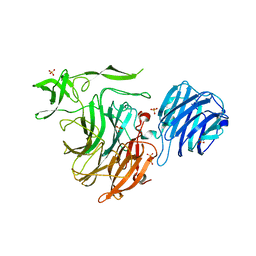 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with N-Acetylneuraminic acid. | | Descriptor: | N-acetyl-alpha-neuraminic acid, SULFATE ION, Sialidase, ... | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YZ3
 
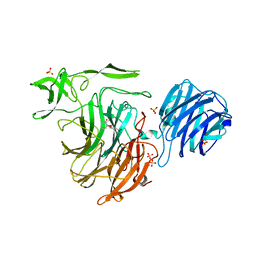 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase, SULFATE ION | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Streptococcus pneumoniae NanC.
To Be Published
|
|
4YZ2
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SULFATE ION, Sialidase NanC | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YZ1
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, apo structure. | | Descriptor: | Putative neuraminidase, SULFATE ION | | Authors: | Lukacik, P, Owen, D.O, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4YZ5
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with 3-Sialyllactose | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, Putative neuraminidase, ... | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
7NW2
 
 | | Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-47 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fearon, D, Douangamath, A, Aimon, A, Brandao-Neto, J, Dias, A, Dunnett, L, Gehrtz, P, Gorrie-Stone, T.J, Lukacik, P, Powell, A.J, Skyner, R, Strain-Damerell, C.M, Zaidman, D, London, N, Walsh, M.A, von Delft, F, Covid Moonshot Consortium | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An automatic pipeline for the design of irreversible derivatives identifies a potent SARS-CoV-2 M pro inhibitor.
Cell Chem Biol, 28, 2021
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
