1HD6
 
 | | PHEROMONE ER-22, NMR | | Descriptor: | PHEROMONE ER-22 | | Authors: | Luginbuhl, P, Liu, A, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 2000-11-09 | | Release date: | 2000-12-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Pheromone Er-22 from Euplotes Raikovi
J.Biomol.NMR, 19, 2001
|
|
1ERY
 
 | | PHEROMONE ER-11, NMR | | Descriptor: | PHEROMONE ER-11 | | Authors: | Luginbuhl, P, Wu, J, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-11 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 5, 1996
|
|
1IM1
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
2VUL
 
 | | Thermostable mutant of ENVIRONMENTALLY ISOLATED GH11 XYLANASE | | Descriptor: | DODECAETHYLENE GLYCOL, GH11 XYLANASE, SULFATE ION | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, Desantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
2VUJ
 
 | | Environmentally isolated GH11 xylanase | | Descriptor: | GH11 XYLANASE, GLYCEROL | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, DeSantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
1E74
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R11E | | Descriptor: | ALPHA-CONOTOXIN IM1(R11E) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E76
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT D5N | | Descriptor: | ALPHA-CONOTOXIN IM1(D5N) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E75
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R7L | | Descriptor: | ALPHA-CONOTOXIN IM1(R7L) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1NH1
 
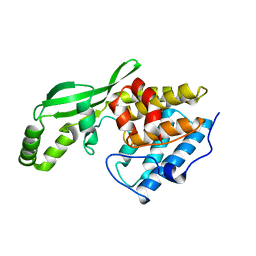 | | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae. | | Descriptor: | Avirulence B protein | | Authors: | Lee, C.C, Wood, M.D, Ng, K, Luginbuhl, P, Spraggon, G, Katagiri, F. | | Deposit date: | 2002-12-18 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae.
Structure, 12, 2004
|
|
1GM0
 
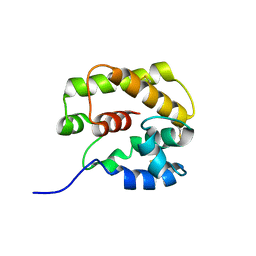 | | A Form of the Pheromone-Binding Protein from Bombyx mori | | Descriptor: | PHEROMONE-BINDING PROTEIN | | Authors: | Horst, R, Damberger, F, Guntert, P, Luginbuhl, P, Nikonova, L, Peng, G, Leal, W.S, Wuthrich, K. | | Deposit date: | 2001-09-05 | | Release date: | 2001-11-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Reveals Novel Intramolecular Regulation Mechanism for Pheromone-Binding and Release
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1ERD
 
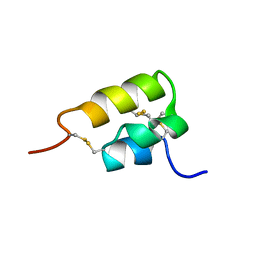 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-2 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-2 | | Authors: | Ottiger, M, Szyperski, T, Luginbuhl, P, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-2 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
1ERC
 
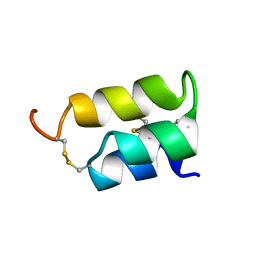 | | THE NMR SOLUTION STRUCTURE OF THE PHEROMONE ER-1 FROM THE CILIATED PROTOZOAN EUPLOTES RAIKOVI | | Descriptor: | PHEROMONE ER-1 | | Authors: | Mronga, S, Luginbuhl, P, Brown, L.R, Ortenzi, C, Luporini, P, Bradshaw, R.A, Wuthrich, K. | | Deposit date: | 1994-02-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the pheromone Er-1 from the ciliated protozoan Euplotes raikovi.
Protein Sci., 3, 1994
|
|
1DC8
 
 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
1DC7
 
 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
