4HHH
 
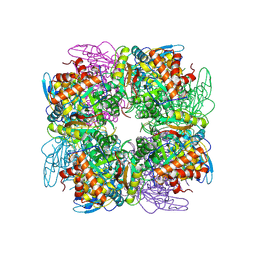 | | Structure of Pisum sativum Rubisco | | Descriptor: | RIBULOSE-1,5-DIPHOSPHATE, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Loewen, P.C, Didychuk, A.L, Switala, J, Loewen, M.C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-31 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Pisum sativum Rubisco with bound ribulose 1,5-bisphosphate.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5KT9
 
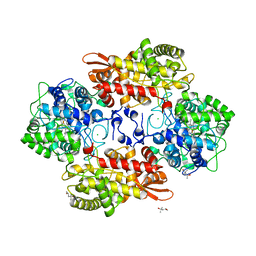 | |
5L02
 
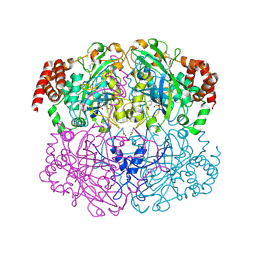
 | | S324T variant of B. pseudomallei KatG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Catalase-peroxidase, PHOSPHATE ION, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the Ser324Thr variant of the catalase-peroxidase (KatG) from Burkholderia pseudomallei
J. Mol. Biol., 345, 2005
|
|
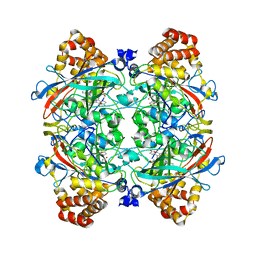
2IQF
 
 | | Crystal structure of Helicobacter pylori catalase compound I | | Descriptor: | ACETATE ION, Catalase, OXYGEN ATOM, ... | | Authors: | Loewen, P.C, Carpena, X, Fita, I. | | Deposit date: | 2006-10-13 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structures and electronic configuration of compound I intermediates of Helicobacter pylori and Penicillium vitale catalases determined by X-ray crystallography and QM/MM density functional theory calculations.
J.Am.Chem.Soc., 129, 2007
|
|
1YE9
 
 | | Crystal structure of proteolytically truncated catalase HPII from E. coli | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, catalase HPII | | Authors: | Loewen, P.C, Chelikani, P, Carpena, X, Fita, I, Perez-Luque, R, Donald, L.J, Switala, J, Duckworth, H.W. | | Deposit date: | 2004-12-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a Large Subunit Catalase Truncated by Proteolytic Cleavage(,)
Biochemistry, 44, 2005
|
|
2A9E
 
 | |
6B9B
 
 | |
6CAW
 
 | |
6CFQ
 
 | |
6CDQ
 
 | |
6CC6
 
 | |
6CEK
 
 | |
4QOM
 
 | | Bacillus pumilus catalase with pyrogallol bound | | Descriptor: | BENZENE-1,2,3-TRIOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOP
 
 | | Structure of Bacillus pumilus catalase with hydroquinone bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOL
 
 | | Structure of Bacillus pumilus catalase | | Descriptor: | ACETATE ION, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOQ
 
 | | Structure of Bacillus pumilus catalase with guaiacol bound | | Descriptor: | CHLORIDE ION, Catalase, Guaiacol, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOO
 
 | | Structure of Bacillus pumilus catalase with resorcinol bound. | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QON
 
 | | Structure of Bacillus pumilus catalase with catechol bound. | | Descriptor: | CATECHOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOR
 
 | | Structure of Bacillus pumilus catalase with chlorophenol bound. | | Descriptor: | 2-CHLOROPHENOL, CHLORIDE ION, Catalase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2014-06-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
5V2D
 
 | |
3TTT
 
 | | Structure of F413Y variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3TTV
 
 | | Structure of the F413E variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3TTU
 
 | | Structure of F413Y/H128N double variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3TTX
 
 | | Structure of the F413K variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
3TTW
 
 | | Structure of the F413E variant of E. coli KatE | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Loewen, P.C, Jha, V. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mutation of Phe413 to Tyr in catalase KatE from Escherichia coli leads to side chain damage and main chain cleavage.
Arch.Biochem.Biophys., 525, 2012
|
|
