8WOQ
 
 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
8WOS
 
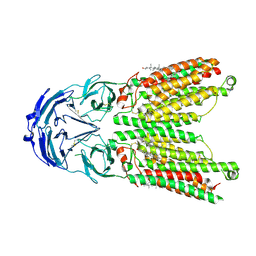 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
5YF8
 
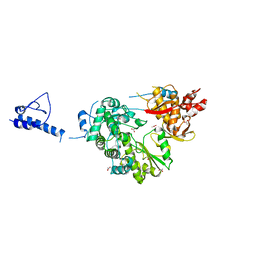 | |
5YF6
 
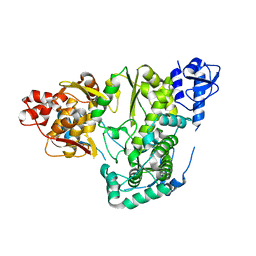 | |
5YF7
 
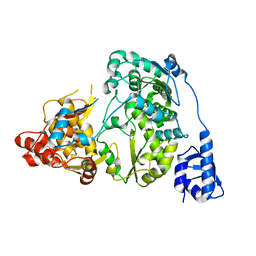 | |
5YF5
 
 | |
6AE4
 
 | |
6AE6
 
 | |
6AE7
 
 | |
6AE5
 
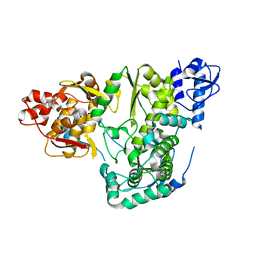 | |
6JJ0
 
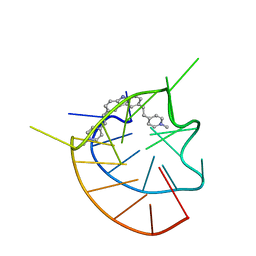 | |
6K63
 
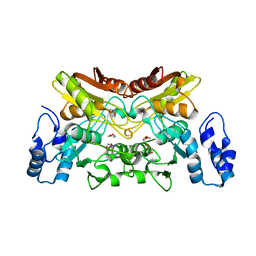 | | The crystal structure of cytidine deaminase from Klebsiella pneumoniae | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Cytidine deaminase, ZINC ION | | Authors: | Liu, W, Shang, F, Lan, J, Chen, Y, Wang, L, Xu, Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Biochemical and structural analysis of the Klebsiella pneumoniae cytidine deaminase CDA.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
7EKJ
 
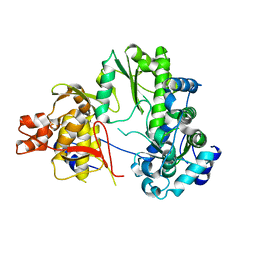 | |
2PW8
 
 | | Crystal structure of sulfo-hirudin complexed to thrombin | | Descriptor: | Hirudin variant-1, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Liu, C.C, Brustad, E, Liu, W, Schultz, P.G. | | Deposit date: | 2007-05-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin.
J.Am.Chem.Soc., 129, 2007
|
|
4UXL
 
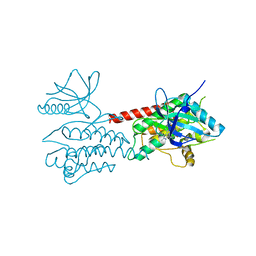 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7LJ3
 
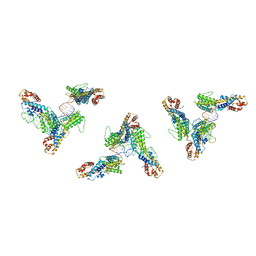 | | Structure of human transfer RNA visualized in the cytomegalovirus, a DNA virus | | Descriptor: | RNA (75-MER), Tegument protein pp150 | | Authors: | Liu, Y.T, Strugatsky, D, Liu, W, Zhou, Z.H. | | Deposit date: | 2021-01-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150.
Nat Commun, 12, 2021
|
|
7LW2
 
 | |
7RFR
 
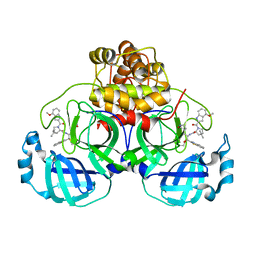 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFU
 
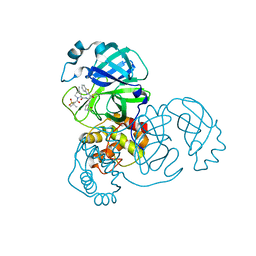 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFW
 
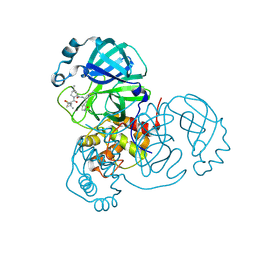 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFS
 
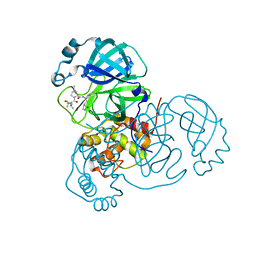 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
5JBM
 
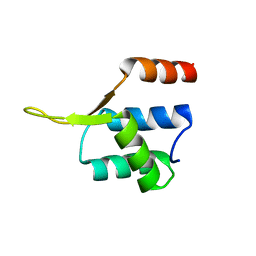 | | Crystal structgure of Cac1 C-terminus | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Churchill, M.E.A, Liu, W, Zhou, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Cac1 subunit of histone chaperone CAF-1 organizes CAF-1-H3/H4 architecture and tetramerizes histones.
Elife, 5, 2016
|
|
8AZU
 
 | | Paired helical tau filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | PHF Tau | | Descriptor: | Microtubule-associated protein tau | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2B
 
 | | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
