3VA6
 
 | |
3VA9
 
 | |
3V9F
 
 | |
5K0X
 
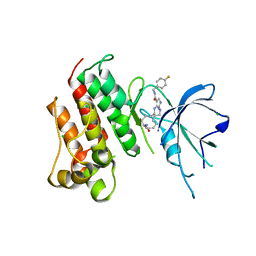 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
2ITG
 
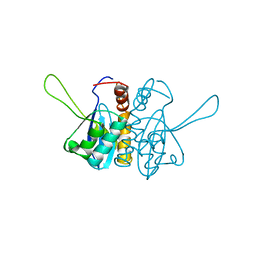 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
4F35
 
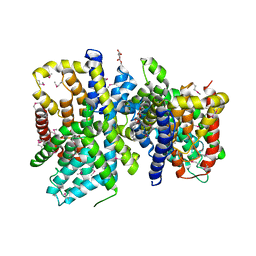 | | Crystal Structure of a bacterial dicarboxylate/sodium symporter | | Descriptor: | CITRIC ACID, SODIUM ION, Transporter, ... | | Authors: | Mancusso, R.L, Gregorio, G.G, Liu, Q, Wang, D.N. | | Deposit date: | 2012-05-08 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter.
Nature, 491, 2012
|
|
8ENA
 
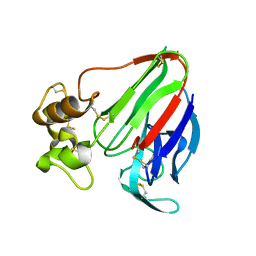 | | Thaumatin native-SAD structure determined at 5 keV with a helium environmet | | Descriptor: | Thaumatin-1 | | Authors: | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
8EN9
 
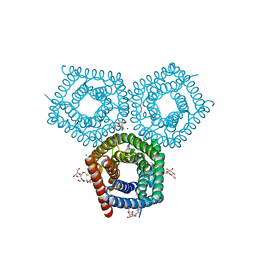 | | TehA native-SAD structure determined at 5 keV with a helium environment | | Descriptor: | CHLORIDE ION, SODIUM ION, Tellurite resistance protein TehA homolog, ... | | Authors: | Karasawa, A, Andi, B, Ruchs, M.R, Shi, W, McSweeney, S, Hendrickson, W.A, Liu, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multi-crystal native-SAD phasing at 5 keV with a helium environment.
Iucrj, 9, 2022
|
|
6ITJ
 
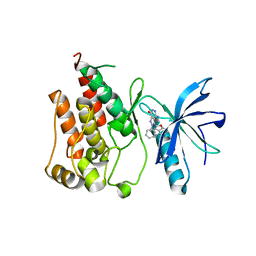 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6IUP
 
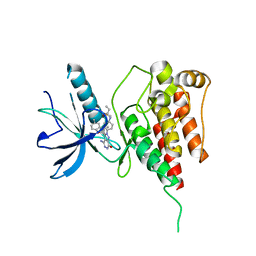 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6NQ8
 
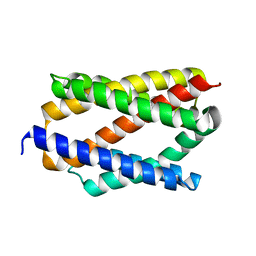 | |
6NQ9
 
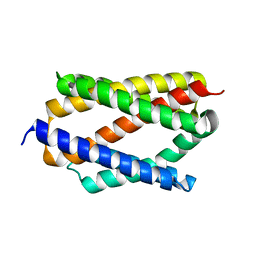 | |
6IUO
 
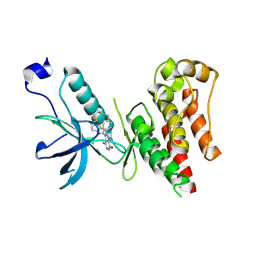 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6NQ7
 
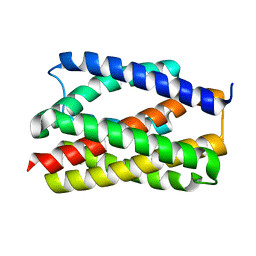 | |
2IMT
 
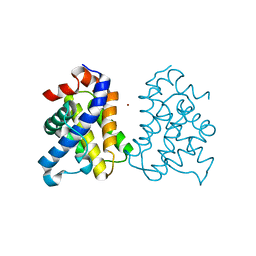 | | The X-ray Structure of a Bak Homodimer Reveals an Inhibitory Zinc Binding Site | | Descriptor: | Apoptosis regulator BAK, ZINC ION | | Authors: | Moldoveanu, T, Liu, Q, Tocilj, A, Watson, M, Shore, G.C, Gehring, K.B. | | Deposit date: | 2006-10-04 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site.
Mol.Cell, 24, 2006
|
|
2IMS
 
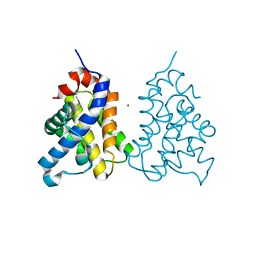 | | The X-ray Structure of a Bak Homodimer Reveals an Inhibitory Zinc Binding Site | | Descriptor: | Apoptosis regulator BAK, ZINC ION | | Authors: | Moldoveanu, T, Liu, Q, Tocilj, A, Watson, M, Shore, G.C, Gehring, K.B. | | Deposit date: | 2006-10-04 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The X-Ray Structure of a BAK Homodimer Reveals an Inhibitory Zinc Binding Site
Mol.Cell, 24, 2006
|
|
5EKE
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
5EKP
 
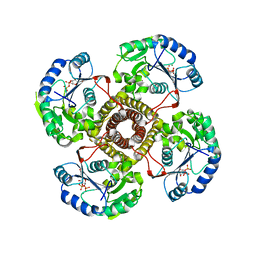 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X6G
 
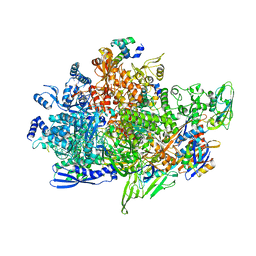 | | Cryo-EM structure of Staphylococcus aureus sigB-dependent RNAP-promoter open complex | | Descriptor: | DNA (70-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
8X6F
 
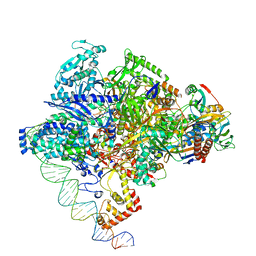 | | Cryo-EM structure of Staphylococcus aureus sigA-dependent RNAP-promoter open complex | | Descriptor: | DNA (71-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
6EGE
 
 | |
3QCQ
 
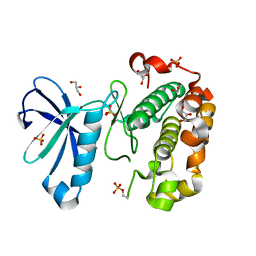 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-(3-Amino-1H-indazol-6-yl)-N4-ethyl-2,4-pyrimidinediamine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-(3-amino-2H-indazol-6-yl)-N~4~-ethylpyrimidine-2,4-diamine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCY
 
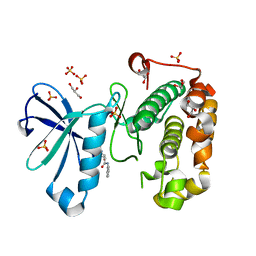 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 4-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-N-phenyl-2-morpholinecarboxamide | | Descriptor: | (2S)-4-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-N-phenylmorpholine-2-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCX
 
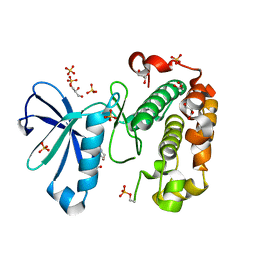 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-{2-Amino-6-[(3R)-3-methyl-4-morpholinyl]-4-pyrimidinyl}-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-{2-amino-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-4-yl}-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
