8GZK
 
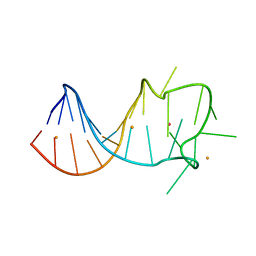 | |
7CRJ
 
 | | Dark State Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5H0R
 
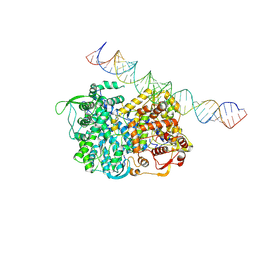 | | RNA dependent RNA polymerase ,vp4,dsRNA | | 分子名称: | RNA (42-MER), RNA-dependent RNA polymerase, VP4 protein | | 著者 | Li, X, Zhou, N, Chen, W, Zhu, B, Wang, X, Xu, B, Wang, J, Liu, H, Cheng, L. | | 登録日 | 2016-10-06 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
7CRI
 
 | | 1 ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2HK9
 
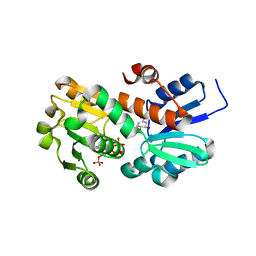 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with shikimate and NADP+ at 2.2 angstrom resolution | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | 登録日 | 2006-07-03 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
3J6Q
 
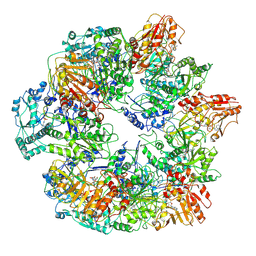 | | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, Structural protein VP3 | | 著者 | Zhu, B, Yang, C, Liu, H, Cheng, L, Song, F, Zeng, S, Huang, X, Ji, G, Zhu, P. | | 登録日 | 2014-03-20 | | 公開日 | 2014-10-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Identification of the active sites in the methyltransferases of a transcribing dsRNA virus.
J.Mol.Biol., 426, 2014
|
|
2HK7
 
 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with mercury at 2.5 angstrom resolution | | 分子名称: | MERCURY (II) ION, Shikimate dehydrogenase | | 著者 | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | 登録日 | 2006-07-03 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
3ZIF
 
 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | 分子名称: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | 著者 | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | 登録日 | 2013-01-09 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
7CRL
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 50 ps after light activation | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRY
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (6.49 mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6KY5
 
 | | Crystal structure of a hydrolase mutant | | 分子名称: | PET hydrolase, SULFATE ION | | 著者 | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | 登録日 | 2019-09-16 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
6AKU
 
 | | Cryo-EM structure of CVA10 empty particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
8GZM
 
 | |
4DWI
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with self complementary DNA, Se-dGTP and Calcium | | 分子名称: | 9-METHYLGUANINE, CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), ... | | 著者 | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | 登録日 | 2012-02-24 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
4DSK
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, PPi and Calcium | | 分子名称: | CALCIUM ION, DNA (5'-D(*GP*GP*CP*TP*AP*CP*AP*GP*GP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*AP*GP*TP*CP*CP*TP*GP*TP*AP*GP*CP*C)-3'), ... | | 著者 | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | 登録日 | 2012-02-19 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
4DSJ
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, dGTP and Calcium | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA, ... | | 著者 | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | 登録日 | 2012-02-19 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
4DSL
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA and Calcium | | 分子名称: | DNA (5'-D(*G*GP*CP*TP*AP*CP*AP*GP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*AP*GP*TP*CP*CP*TP*GP*TP*AP*GP*CP*C)-3'), DNA polymerase, ... | | 著者 | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | 登録日 | 2012-02-19 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
4DSI
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA, Se-dGTP and Calcium | | 分子名称: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA, ... | | 著者 | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | 登録日 | 2012-02-19 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | 分子名称: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | 著者 | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | 登録日 | 2010-03-24 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
5Y1Z
 
 | | Crystal structure of ZMYND8 PHD-BROMO-PWWP tandem in complex with Drebrin ADF-H domain | | 分子名称: | Drebrin, GLYCEROL, Protein kinase C-binding protein 1, ... | | 著者 | Yao, N, Li, J, Liu, H, Wan, J, Liu, W, Zhang, M. | | 登録日 | 2017-07-22 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.676 Å) | | 主引用文献 | The Structure of the ZMYND8/Drebrin Complex Suggests a Cytoplasmic Sequestering Mechanism of ZMYND8 by Drebrin
Structure, 25, 2017
|
|
4EMM
 
 | |
