6PB4
 
 | |
6PB5
 
 | |
6PB6
 
 | |
6XQB
 
 | | SARS-CoV-2 RdRp/RNA complex | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
8U3B
 
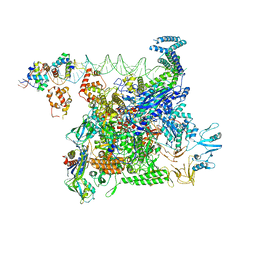 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
6PMJ
 
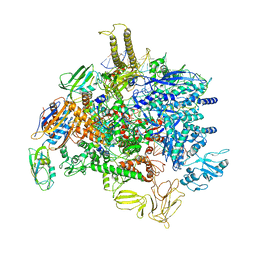 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
4S20
 
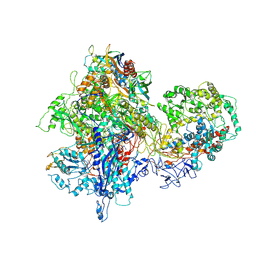 | | Structural basis for transcription reactivation by RapA | | Descriptor: | 5'-D(P*AP*CP*GP*AP*CP*TP*GP*AP*GP*CP*CP*GP*AP*TP*G)-3', 5'-R(P*AP*UP*CP*GP*GP*CP*UP*CP*A)-3', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, B, Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-01-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Structural basis for transcription reactivation by RapA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6PMI
 
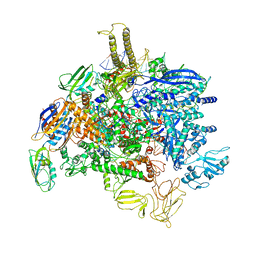 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
7AMT
 
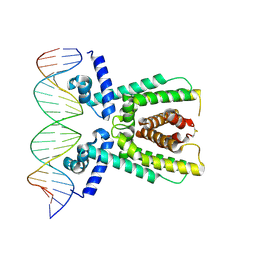 | | Structure of LuxR with DNA (activation) | | Descriptor: | DNA (5'-D(P*AP*TP*AP*AP*TP*GP*AP*CP*AP*TP*TP*AP*CP*TP*GP*TP*AP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*AP*GP*TP*AP*AP*TP*GP*TP*CP*AP*TP*TP*AP*T)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
8DKC
 
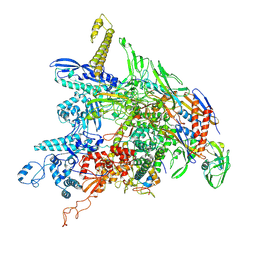 | | P. gingivalis RNA Polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Bu, F. | | Deposit date: | 2022-07-05 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure of Porphyromonas gingivalis RNA Polymerase.
J.Mol.Biol., 436, 2024
|
|
6XZ1
 
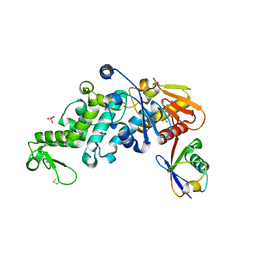 | | Conjugate of the HECT domain of HUWE1 with ubiquitin | | Descriptor: | HECT, UBA and WWE domain containing 1, isoform CRA_a, ... | | Authors: | Liu, B, Seenivasan, A, Nair, R, Chen, D, Lowe, E.D, Lorenz, S. | | Deposit date: | 2020-01-31 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution and Structural Analysis of a HECT Ligase-Ubiquitin Complex via an Activity-Based Probe.
Acs Chem.Biol., 16, 2021
|
|
2BD5
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Lys-Ser at pH 5 and immersed in pH 9 buffer for 30 seconds | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
7AMN
 
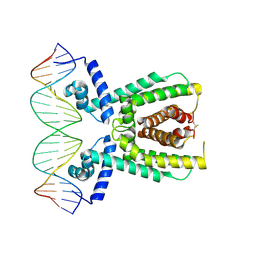 | | Structure of LuxR with DNA (repression) | | Descriptor: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
2BD3
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Lys-Ala-NH2 at pH 5.0 | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BD4
 
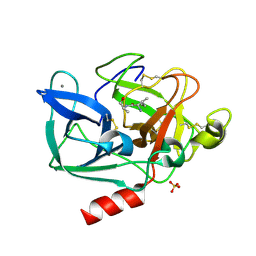 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Lys-Ser at pH 5.0 | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BD9
 
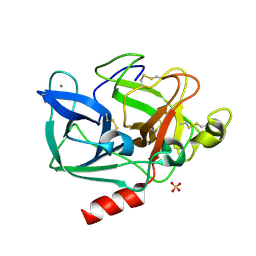 | |
2BD8
 
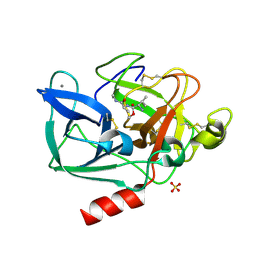 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 (50 min soak) and immersed in pH 9 buffer for 30 seconds | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BDA
 
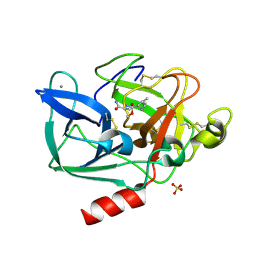 | | Porcine pancreatic elastase complexed with N-acetyl-NPI and Ala-Ala at pH 5.0 | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, NPI, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BDC
 
 | |
2BDB
 
 | | Porcine pancreatic elastase complexed with Asn-Pro-Ile and Ala-Ala at pH 5.0 | | Descriptor: | ALANINE, CALCIUM ION, Elastase-1, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BD2
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BD7
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 (50 min soak) | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
2BB4
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Asp-Phe at pH 5.0 | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, SULFATE ION, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-17 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
7YG7
 
 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
