6M6R
 
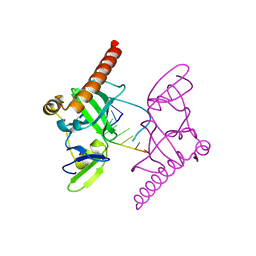 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 8-mer ssRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*CP*GP*CP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6Q
 
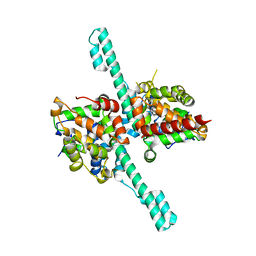 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) N-terminal domain | | Descriptor: | Dicer Related Helicase | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6S
 
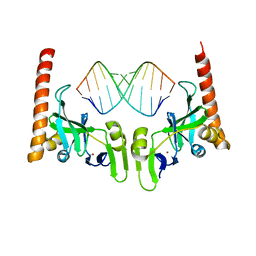 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 12-mer dsRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
7QDV
 
 | |
5A2U
 
 | |
6JWN
 
 | |
5IG9
 
 | |
6JWM
 
 | |
4YLH
 
 | | Crystal structure of DpgC with bound substrate analog and Xe on oxygen diffusion pathway | | Descriptor: | DpgC, XENON, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Li, K, Di Russo, N.V, Condurso, H.L, Roitberg, A.E, Bruner, S.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Oxygen diffusion pathways in a cofactor-independent dioxygenase.
Chem Sci, 6, 2015
|
|
5IG8
 
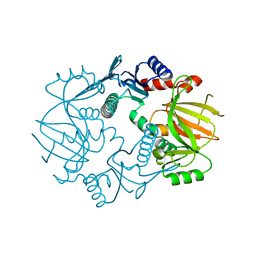 | |
4YHB
 
 | | Crystal structure of a siderophore utilization protein from T. fusca | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Iron-chelator utilization protein, ... | | Authors: | Li, K, Bruner, S.D. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8892 Å) | | Cite: | Structure and Mechanism of the Siderophore-Interacting Protein from the Fuscachelin Gene Cluster of Thermobifida fusca.
Biochemistry, 54, 2015
|
|
5GPJ
 
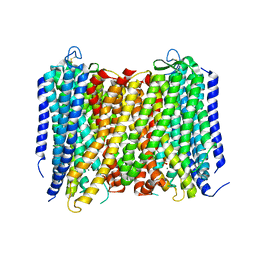 | | Crystal Structure of Proton-Pumping Pyrophosphatase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Pyrophosphate-energized vacuolar membrane proton pump | | Authors: | Li, K.M, Tsai, J.Y, Sun, Y.J. | | Deposit date: | 2016-08-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism
Nat Commun, 7, 2016
|
|
6HPN
 
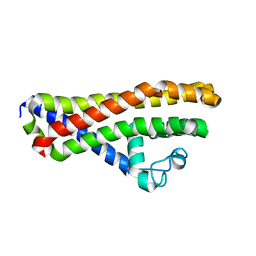 | | Crystal structure of Borrelia spielmanii WP_012665240 (BSA64) - an orthologous protein to B. burgdorferi BBA64 | | Descriptor: | Antigen, P35 | | Authors: | Brangulis, K, Akopjana, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the outer surface proteins from Borrelia burgdorferi paralogous gene family 54 that are thought to be the key players in the pathogenesis of Lyme disease.
J.Struct.Biol., 210, 2020
|
|
6RJW
 
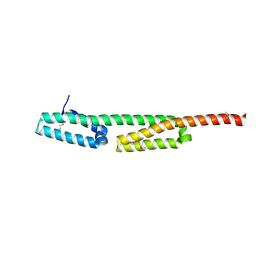 | |
6RJX
 
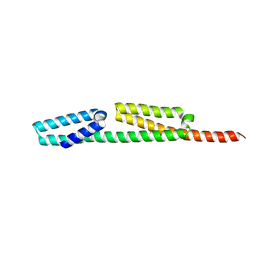 | |
6ROC
 
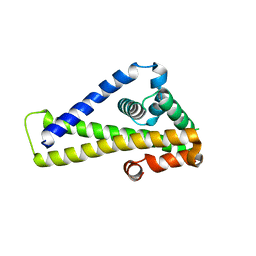 | | Crystal structure of Borrelia burgdorferi outer surface protein BBA69, mutant Leu214Met (Se-Met data) | | Descriptor: | Putative surface protein | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2019-05-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Borrelia burgdorferi outer surface protein BBA69 in comparison to the paralogous protein CspA.
Ticks Tick Borne Dis, 10, 2019
|
|
1HP2
 
 | | SOLUTION STRUCTURE OF A TOXIN FROM THE SCORPION TITYUS SERRULATUS (TSTX-K ALPHA) DETERMINED BY NMR. | | Descriptor: | TITYUSTOXIN K ALPHA | | Authors: | Ellis, K.C, Tenenholz, T.C, Gilly, W.F, Blaustein, M.P, Weber, D.J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-06-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Interaction of a toxin from the scorpion Tityus serrulatus with a cloned K+ channel from squid (sqKv1A).
Biochemistry, 40, 2001
|
|
2QIY
 
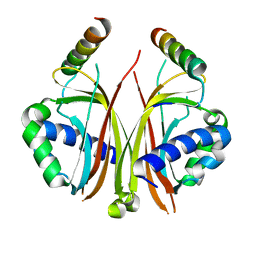 | |
4BG0
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Baumanis, V, Tars, K. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
4CBE
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi (Native) | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-10-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
1ZX2
 
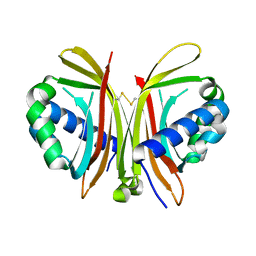 | | Crystal Structure of Yeast UBP3-associated Protein BRE5 | | Descriptor: | UBP3-associated protein BRE5 | | Authors: | Li, K, Zhao, K, Ossareh-Nazari, B, Da, G, Dargemont, C, Marmorstein, R. | | Deposit date: | 2005-06-06 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor
J.Biol.Chem., 280, 2005
|
|
4BG5
 
 | |
4ALY
 
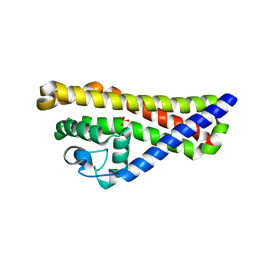 | | Borrelia burgdorferi outer surface lipoprotein BBA64 | | Descriptor: | P35 ANTIGEN, SULFATE ION | | Authors: | Brangulis, K, Tars, K, Petrovskis, I, Kazaks, A, Ranka, R, Baumanis, V. | | Deposit date: | 2012-03-06 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an Outer Surface Lipoprotein Bba64 from the Lyme Disease Agent Borrelia Burgdorferi which is Critical to Ensure Infection After a Tick Bite
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AXZ
 
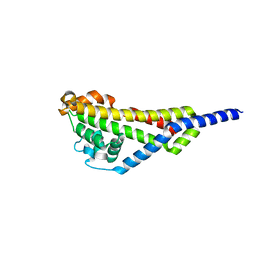 | | Borrelia burgdorferi outer surface lipoprotein BBA73 | | Descriptor: | PUTATIVE ANTIGEN P35 | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Baumanis, V, Tars, K. | | Deposit date: | 2012-06-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Characterization of the Borrelia Burgdorferi Outer Surface Protein Bba73 Implicates Dimerization as a Functional Mechanism.
Biochem.Biophys.Res.Commun., 434, 2013
|
|
4BOD
 
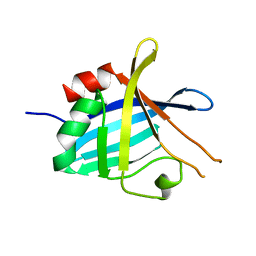 | | Complement regulator acquiring outer surface protein BbCRASP-4 or ErpC from Borrelia burgdorferi | | Descriptor: | ERPC | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Baumanis, V, Tars, K. | | Deposit date: | 2013-05-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of the Erp Protein Family Members Erpp and Erpc from Borrelia Burgdorferi Reveal the Reason for Different Affinities for Complement Regulator Factor H.
Biochim.Biophys.Acta, 1854, 2015
|
|
