5DMM
 
 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Metallated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, BETA-MERCAPTOETHANOL, Homocysteine S-methyltransferase, ... | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
6JWN
 
 | |
6KEY
 
 | |
5IG8
 
 | |
5DML
 
 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Oxidized form | | Descriptor: | CHLORIDE ION, Homocysteine S-methyltransferase | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
6M6R
 
 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 8-mer ssRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*CP*GP*CP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6Q
 
 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) N-terminal domain | | Descriptor: | Dicer Related Helicase | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
6M6S
 
 | | Crystal structure of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3) C-terminal domain with 5'-ppp 12-mer dsRNA | | Descriptor: | Dicer Related Helicase, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, K, Zheng, J, Wirawan, M, Xiong, Z, Fedorova, O, Griffin, P, Plyle, A, Luo, D. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the structure and RNA-binding specificity of Caenorhabditis elegans Dicer-related helicase 3 (DRH-3).
Nucleic Acids Res., 49, 2021
|
|
5GPJ
 
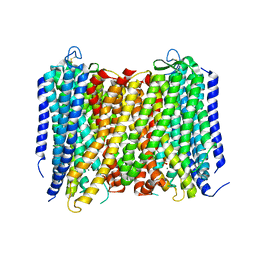 | | Crystal Structure of Proton-Pumping Pyrophosphatase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Pyrophosphate-energized vacuolar membrane proton pump | | Authors: | Li, K.M, Tsai, J.Y, Sun, Y.J. | | Deposit date: | 2016-08-03 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism
Nat Commun, 7, 2016
|
|
5DMN
 
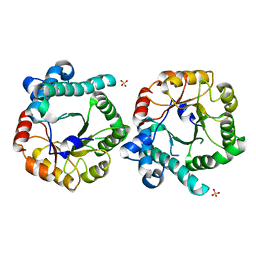 | | Crystal Structure of the Homocysteine Methyltransferase MmuM from Escherichia coli, Apo form | | Descriptor: | Homocysteine S-methyltransferase, SULFATE ION | | Authors: | Li, K, Li, G, Bradbury, L.M.T, Andrew, H.D, Bruner, S.D. | | Deposit date: | 2015-09-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | Crystal structure of the homocysteine methyltransferase MmuM from Escherichia coli.
Biochem.J., 473, 2016
|
|
6JWM
 
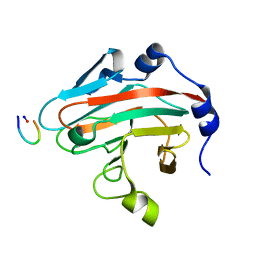 | |
6V2U
 
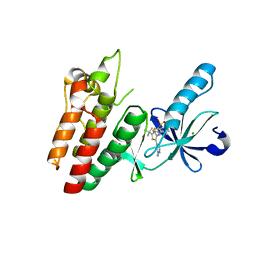 | |
6V2W
 
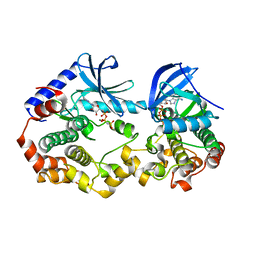 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Park, E, Eck, M.J. | | Deposit date: | 2019-11-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5V6J
 
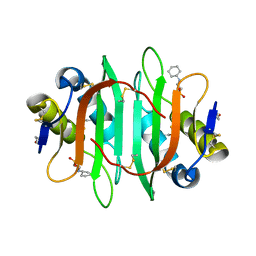 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TMV resistance protein Y3 | | Authors: | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V6I
 
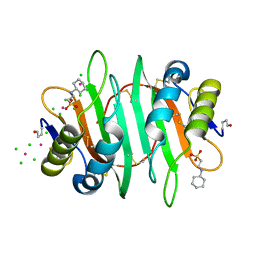 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity - Pt derivative | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, PLATINUM (II) ION, ... | | Authors: | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-16 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DH1
 
 | |
4BG0
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Baumanis, V, Tars, K. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
4CBE
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi (Native) | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-10-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
1HP2
 
 | | SOLUTION STRUCTURE OF A TOXIN FROM THE SCORPION TITYUS SERRULATUS (TSTX-K ALPHA) DETERMINED BY NMR. | | Descriptor: | TITYUSTOXIN K ALPHA | | Authors: | Ellis, K.C, Tenenholz, T.C, Gilly, W.F, Blaustein, M.P, Weber, D.J. | | Deposit date: | 2000-12-12 | | Release date: | 2001-06-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Interaction of a toxin from the scorpion Tityus serrulatus with a cloned K+ channel from squid (sqKv1A).
Biochemistry, 40, 2001
|
|
4D53
 
 | | Outer surface protein BB0689 from Borrelia burgdorferi | | Descriptor: | BB0689 | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2014-11-02 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Analysis of Bb0689 from Borrelia Burgdorferi, a Member of the Bacterial CAP Superfamily.
J.Struct.Biol., 192, 2015
|
|
4B2F
 
 | |
4BG5
 
 | |
4BOD
 
 | | Complement regulator acquiring outer surface protein BbCRASP-4 or ErpC from Borrelia burgdorferi | | Descriptor: | ERPC | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Baumanis, V, Tars, K. | | Deposit date: | 2013-05-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of the Erp Protein Family Members Erpp and Erpc from Borrelia Burgdorferi Reveal the Reason for Different Affinities for Complement Regulator Factor H.
Biochim.Biophys.Acta, 1854, 2015
|
|
4BXM
 
 | | Complement regulator acquiring outer surface protein BbCRASP-4 or ErpC from Borrelia burgdorferi | | Descriptor: | ERPC | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-07-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Erp Protein Family Members Erpp and Erpc from Borrelia Burgdorferi Reveal the Reason for Different Affinities for Complement Regulator Factor H.
Biochim.Biophys.Acta, 1854, 2015
|
|
4AXZ
 
 | | Borrelia burgdorferi outer surface lipoprotein BBA73 | | Descriptor: | PUTATIVE ANTIGEN P35 | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Baumanis, V, Tars, K. | | Deposit date: | 2012-06-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Characterization of the Borrelia Burgdorferi Outer Surface Protein Bba73 Implicates Dimerization as a Functional Mechanism.
Biochem.Biophys.Res.Commun., 434, 2013
|
|
