4KPR
 
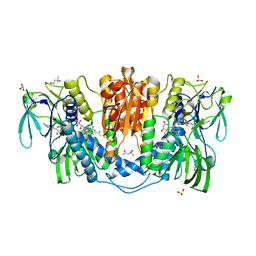 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
3IHG
 
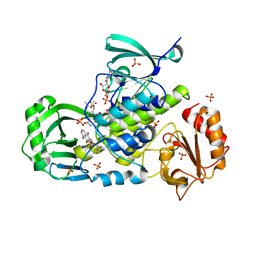 | | Crystal structure of a ternary complex of aklavinone-11 hydroxylase with FAD and aklavinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RdmE, SULFATE ION, ... | | Authors: | Lindqvist, Y, Koskiniemi, H, Jansson, A, Sandalova, T, Schneider, G. | | Deposit date: | 2009-07-30 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis.
J.Mol.Biol., 393, 2009
|
|
1GOX
 
 | |
1TRK
 
 | |
1GYL
 
 | |
1RPA
 
 | |
1RPT
 
 | |
1QHW
 
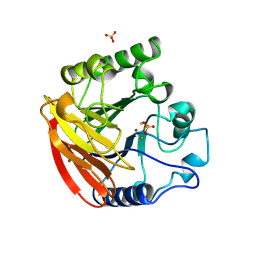 | | PURPLE ACID PHOSPHATASE FROM RAT BONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PROTEIN (PURPLE ACID PHOSPHATASE), ... | | Authors: | Lindqvist, Y, Johansson, E, Kaija, H, Vihko, P, Schneider, G. | | Deposit date: | 1999-03-26 | | Release date: | 1999-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a mammalian purple acid phosphatase at 2.2 A resolution with a mu-(hydr)oxo bridged di-iron center.
J.Mol.Biol., 291, 1999
|
|
1AFR
 
 | |
1VGQ
 
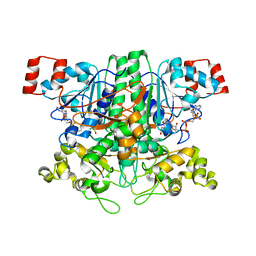 | | Formyl-CoA transferase mutant Asp169 to Ala | | Descriptor: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
3LCP
 
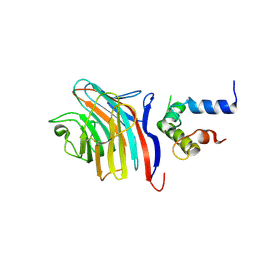 | | Crystal structure of the carbohydrate recognition domain of LMAN1 in complex with MCFD2 | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53 | | Authors: | Wigren, E, Bourhis, J.M, Kursula, I, Guy, J.E, Lindqvist, Y. | | Deposit date: | 2010-01-11 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the LMAN1-CRD/MCFD2 transport receptor complex provides insight into combined deficiency of factor V and factor VIII.
Febs Lett., 584, 2010
|
|
2XZ1
 
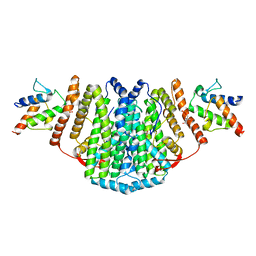 | | The Structure of the 2:2 (Fully Occupied) Complex Between Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) and Acyl Carrier Protein. | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, ACYL CARRIER PROTEIN 1, CHLOROPLASTIC, ... | | Authors: | Guy, J.E, Moche, M, Whittle, E, Lengqvist, J, Shanklin, J, Lindqvist, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Remote Control of Regioselectivity in Acyl-Acyl Carrier Protein-Desaturases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XZ0
 
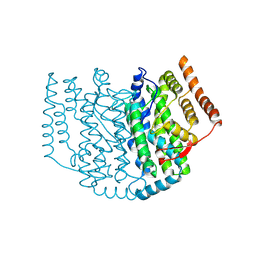 | | The Structure of the 2:1 (Partially Occupied) Complex Between Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) and Acyl Carrier Protein. | | Descriptor: | 1,2-ETHANEDIOL, ACYL CARRIER PROTEIN 1, CHLOROPLASTIC, ... | | Authors: | Moche, M, Guy, J.E, Whittle, E, Lengqvist, J, Shanklin, J, Lindqvist, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Remote Control of Regioselectivity in Acyl-Acyl Carrier Protein-Desaturases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 4 | | Descriptor: | ((1RS,3SR)-2,2-DICHLORO-N-[(R)-1-(4-CHLOROPHENYL)ETHYL]-1-ETHYL-3-METHYLCYCLOPROPANECARBOXAMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
4V0J
 
 | | The channel-block Ser202Glu, Thr104Lys double mutant of Stearoyl-ACP- Desaturase from Castor bean (Ricinus communis) | | Descriptor: | ACYL-[ACYL-CARRIER-PROTEIN] DESATURASE, CHLOROPLASTIC, FE (II) ION, ... | | Authors: | Moche, M, Guy, J, Whittle, E, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2014-09-17 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Half-of-the-Sites Reactivity of the Castor Delta9-18:0-Acp Desaturase.
Plant Physiol., 169, 2015
|
|
7PP7
 
 | | Thunberia alata 16:0-ACP desaturase | | Descriptor: | Acyl-[acyl-carrier-protein] 6-desaturase, FE (III) ION | | Authors: | Guy, J.E, Whittle, E, Cai, Y, Chai, J, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2021-09-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselectivity mechanism of the Thunbergia alata Delta 6-16:0-acyl carrier protein desaturase.
Plant Physiol., 188, 2022
|
|
5RUB
 
 | | CRYSTALLOGRAPHIC REFINEMENT AND STRUCTURE OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE FROM RHODOSPIRILLUM RUBRUM AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Schneider, G, Lindqvist, Y, Lundqvist, T. | | Deposit date: | 1990-05-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic refinement and structure of ribulose-1,5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 A resolution.
J.Mol.Biol., 211, 1990
|
|
2CND
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-DEPENDENT NITRATE REDUCTASE | | Authors: | Lu, G, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-02-01 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
4STD
 
 | | HIGH RESOLUTION STRUCTURES OF SCYTALONE DEHYDRATASE-INHIBITOR COMPLEXES CRYSTALLIZED AT PHYSIOLOGICAL PH | | Descriptor: | N-[1-(4-BROMOPHENYL)ETHYL]-5-FLUORO SALICYLAMIDE, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
4HU2
 
 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain A and B | | Descriptor: | PROBABLE CONSERVED LIPOPROTEIN LPPS, SULFATE ION | | Authors: | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HUC
 
 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain B and C | | Descriptor: | ACETATE ION, PROBABLE CONSERVED LIPOPROTEIN LPPS, SODIUM ION | | Authors: | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3EAN
 
 | | Crystal structure of recombinant rat selenoprotein thioredoxin reductase 1 with reduced C-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Sandalova, T, Cheng, Q, Lindqvist, Y, Arner, E. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1.
J.Biol.Chem., 284, 2009
|
|
2C31
 
 | | CRYSTAL STRUCTURE OF OXALYL-COA DECARBOXYLASE IN COMPLEX WITH THE COFACTOR DERIVATIVE THIAMIN-2-THIAZOLONE DIPHOSPHATE AND ADENOSINE DIPHOSPHATE | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Berthold, C.L, Moussatche, P, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2005-10-03 | | Release date: | 2005-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for Activation of the Thiamin Diphosphate-Dependent Enzyme Oxalyl-Coa Decarboxylase by Adenosine Diphosphate.
J.Biol.Chem., 280, 2005
|
|
3EAO
 
 | | Crystal structure of recombinant rat selenoprotein thioredoxin reductase 1 with oxidized C-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1, ... | | Authors: | Sandalova, T, Cheng, Q, Lindqvist, Y, Arner, E. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1.
J.Biol.Chem., 284, 2009
|
|
5STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 2 | | Descriptor: | (6,7-DIFLUORO-QUINAZOLIN-4-YL)-(1-METHYL-2,2-DIPHENYL-ETHYL)-AMINE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
