8JBQ
 
 | |
2MGX
 
 | | NMR structure of SRA1p C-terminal domain | | Descriptor: | Steroid receptor RNA activator 1 | | Authors: | Bilinovich, S.M, Davis, C.M, Morris, D.L, Ray, L.A, Prokop, J.W, Buchan, G.J, Leeper, T.C. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domain of SRA1p Has a Fold More Similar to PRP18 than to an RRM and Does Not Directly Bind to the SRA1 RNA STR7 Region.
J.Mol.Biol., 426, 2014
|
|
2MZC
 
 | | Metal Binding of Glutaredoxins | | Descriptor: | Glutaredoxin, SILVER ION | | Authors: | Bilinovich, S.M, Caporoso, J.A, Taraboletti, A, Duangjumpa, N, Panzner, M.J, Prokop, J.W, Shriver, L.P, Leeper, T.C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal Binding of Glutaredoxins
To be Published
|
|
6LNN
 
 | |
6LZ6
 
 | |
6LZ8
 
 | |
8JC7
 
 | | Cryo-EM structure of Vibrio campbellii alpha-hemolysin | | Descriptor: | CALCIUM ION, Hemolysin, POTASSIUM ION | | Authors: | Wang, C.H, Yeh, M.K, Ho, M.C, Lin, S.M. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | Structural basis for calcium-stimulating pore formation of Vibrio alpha-hemolysin.
Nat Commun, 14, 2023
|
|
6KL5
 
 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | Descriptor: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL6
 
 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | Descriptor: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | Authors: | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL2
 
 | |
8XP9
 
 | | Crystal structure of d(ACGCCGT/ACGGCGT) | | Descriptor: | DNA (5'-D(P*AP*CP*GP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3') | | Authors: | Hou, M.H, Lin, S.M, Lin, Y.J, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
7V9R
 
 | | Crystal Structure of the heptameric EcHsp60 | | Descriptor: | 60 kDa chaperonin | | Authors: | Lai, M.C, Lin, S.M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of dimeric and heptameric mtHsp60 reveal the mechanism of chaperonin inactivation.
Life Sci Alliance, 6, 2023
|
|
7V98
 
 | | Crystal Structure of the Dimeric EcHsp60 | | Descriptor: | 60 kDa chaperonin | | Authors: | Lai, M.C, Lin, S.M. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of dimeric and heptameric mtHsp60 reveal the mechanism of chaperonin inactivation.
Life Sci Alliance, 6, 2023
|
|
8WNB
 
 | | Crystal structure of d(ACGCCGT/ACGICGT) | | Descriptor: | DNA (5'-D(P*AP*CP*GP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*IP*CP*GP*T)-3') | | Authors: | Hou, M.H, Huang, H.T, Lin, S.M. | | Deposit date: | 2023-10-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
8XPB
 
 | | Crystal structure of d(ACGCCGT/ACGGCGT) in complex with Echinomycin | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Huang, H.T, Lin, S.M, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
8XP8
 
 | | Crystal structure of d(ACGmCCGT/ACGGCGT) in complex with Echinomycin | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*(5CM)P*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Lin, S.M, Neidle, H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
8XPA
 
 | | Crystal structure of d(ACGmCCGT/ACGGCGT) | | Descriptor: | DNA (5'-D(P*AP*CP*GP*(5CM)P*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Hou, M.H, Lin, S.M, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
7DYD
 
 | |
6L76
 
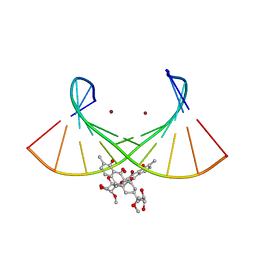 | | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Jhan, C.R, Satange, R.B, Lin, S.M. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
6XRA
 
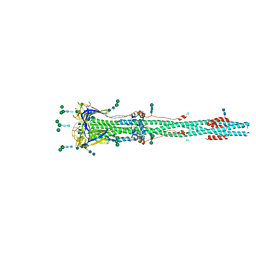 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Rits-Volloch, S, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6XR8
 
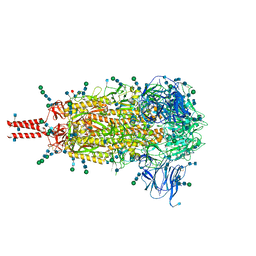 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
3UGE
 
 | | Silver Metallated Pseudomonas aeruginosa Azurin at 1.70 A | | Descriptor: | Azurin, SILVER ION | | Authors: | Panzner, M.J, Billinovich, S.M, Parker, J.A, Bladholm, E, Berry, S.M, Ziegler, C.J, Leeper, T.C. | | Deposit date: | 2011-11-02 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Silver Metallation of Pseudomonas aeruginosa Azurin
To be Published
|
|
7N1W
 
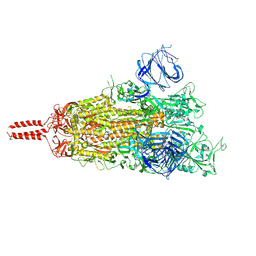 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1Y
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1U
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
