8OLN
 
 | | DI1 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLQ
 
 | | DI3 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL2
 
 | | Murine type II Abeta fibril from APP23 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL5
 
 | | Murine type II Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL7
 
 | | MurineArc type I Abeta fibril from tg-APPArcSwe mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLG
 
 | | DI2 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLO
 
 | | Murine type III Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
6UIE
 
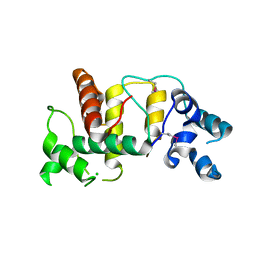 | | Structure of the cytoplasmic domain of the T3SS sorting platform protein PscK from P. aeruginosa | | Descriptor: | CHLORIDE ION, Type III export protein PscK | | Authors: | Muthuramalingam, M, Lovell, S, Battaile, K.P, Picking, W.D. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structures of SctK and SctD from Pseudomonas aeruginosa Reveal the Interface of the Type III Secretion System Basal Body and Sorting Platform.
J.Mol.Biol., 432, 2020
|
|
5G5W
 
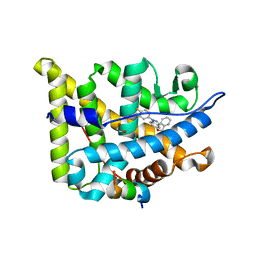 | | Structure guided design and discovery of Indazole ethers as highly potent, non-steroidal Glucocorticoid receptor modulators | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-trifluoro-N-[(1R,2S)-1-{[1-(4-fluorophenyl)-1H-indazol-5-yl]oxy}-1-phenylpropan-2-yl]acetamide, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Hemmerling, M, Edman, K, Lepisto, M, Eriksson, A, Ivanova, S, Dahmen, J, Rehwinkel, H, Berger, M, Hendrickx, R, Dearman, M, Jellesmark-Jensen, T, Wissler, L, Hansson, T. | | Deposit date: | 2016-06-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Indazole Ethers as Novel, Potent, Non-Steroidal Glucocorticoid Receptor Modulators.
Bioorg.Med.Chem.Lett., 26, 2017
|
|
8BSE
 
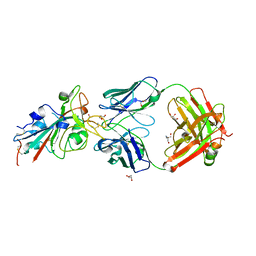 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD) in complex with 1D1 Fab | | Descriptor: | 1D1 FAB HEAVY CHAIN, 1D1 FAB LIGHT CHAIN, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8BSF
 
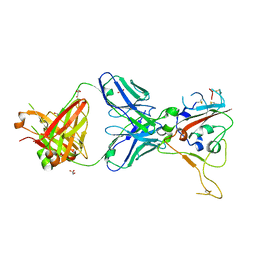 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD-beta variant) in complex with 3D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3D2 FAB HEAVY CHAIN, 3D2 FAB LIGHT CHAIN, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8QG7
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl-pent-4-ynyl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGE
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-pent-4-ynyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGJ
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-4-carbamimidamido-butanamide | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGI
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-(phenylmethyl)urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGL
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-5-[[[1,1-bis(oxidanyl)-1$l^{6}-thia-3-azabicyclo[1.1.0]butan-1-yl]amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To be published
|
|
8QGA
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-naphthalen-1-yl-urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QG8
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]propyl]guanidine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGN
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To be published
|
|
8QGK
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]propyl]ethanamide | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QG9
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl-propan-2-yl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGH
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl]amino]propyl]-3-[(azanylidene-$l^{4}-azanylidene)amino]propanamide | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QG2
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-2-(2-azanyl-2-oxidanylidene-ethyl)benzamide | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QG4
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGM
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-[(dimethylsulfamoylamino)methyl]-3,4-bis(oxidanyl)oxolan-2-yl]purine, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
