1I1F
 
 | | Crystal structure of human class i mhc (hla-a2.1) complexed with beta 2-microglobulin and hiv-rt variant peptide i1y | | Descriptor: | PROTEIN (BETA 2-MICROGLOBULIN), PROTEIN (CLASS I HISTOCOMPATIBILITY ANTIGEN, GOGO-A0201 ALPHA CHAIN), ... | | Authors: | Kirksey, T.J, Pogue-Caley, R.R, Frelinger, J.A, Collins, E.J. | | Deposit date: | 1999-04-16 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for the increased immunogenicity of two HIV-reverse transcriptase peptide variant/class I major histocompatibility complexes.
J.Biol.Chem., 274, 1999
|
|
1M2R
 
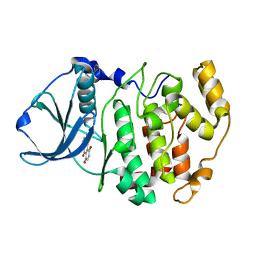 | | Crystal structure of 5,8-di-amino-1,4-di-hydroxy-anthraquinone/CK2 kinase complex | | Descriptor: | 5,8-DI-AMINO-1,4-DIHYDROXY-ANTHRAQUINONE, CASEIN kinase II, alpha chain | | Authors: | De Moliner, E, Moro, S, Sarno, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
3RWO
 
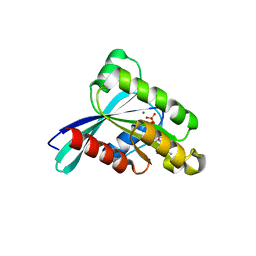 | | Crystal structure of YPT32 in complex with GDP | | Descriptor: | GTP-binding protein YPT32/YPT11, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sultana, A, Jin, Y, Dregger, C, Franklin, E, Weisman, L.S, Khan, A.R. | | Deposit date: | 2011-05-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The activation cycle of Rab GTPase Ypt32 reveals structural determinants of effector recruitment and GDI binding.
Febs Lett., 585, 2011
|
|
1M2P
 
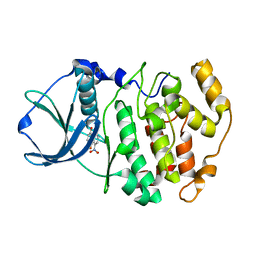 | | Crystal structure of 1,8-di-hydroxy-4-nitro-anthraquinone/CK2 kinase complex | | Descriptor: | 1,8-DI-HYDROXY-4-NITRO-ANTHRAQUINONE, Casein kinase II, alpha chain | | Authors: | De Moliner, E, Moro, S, Sarno, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
2G3K
 
 | | Crystal structure of the C-terminal domain of Vps28 | | Descriptor: | Vacuolar protein sorting-associated protein VPS28 | | Authors: | Pineda-Molina, E, Belrhali, H, Piefer, A.J, Akula, I, Bates, P, Weissenhorn, W. | | Deposit date: | 2006-02-20 | | Release date: | 2006-06-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment.
Traffic, 7, 2006
|
|
2XX9
 
 | | Crystal structure of 1-((2-fluoro-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX7
 
 | | Crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | 1-[4-(1-PYRROLIDINYLCARBONYL)PHENYL]-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XXH
 
 | | Crystal structure of 1-(4-(2-oxo-2-(1-pyrrolidinyl)ethyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.5A resolution. | | Descriptor: | 1-{4-[2-OXO-2-(1-PYRROLIDINYL)ETHYL]PHENYL}-3-( TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX8
 
 | | Crystal structure of N,N-dimethyl-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)benzamide in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
1M2Q
 
 | | Crystal structure of 1,8-di-hydroxy-4-nitro-xanten-9-one/CK2 kinase complex | | Descriptor: | 1,8-DI-HYDROXY-4-NITRO-XANTHEN-9-ONE, Casein kinase II, alpha chain | | Authors: | De Moliner, E, Sarno, S, Moro, S, Zagotto, G, Zanotti, G, Pinna, L.A, Battistutta, R. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibition of protein kinase CK2 by anthraquinone-related compounds. A
structural insight
J.Biol.Chem., 278, 2003
|
|
2XXI
 
 | | Crystal structure of 1-((4-(3-(trifluoromethyl)-6,7-dihydropyrano(4,3- c(pyrazol-1(4H)-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.6A resolution. | | Descriptor: | 1-({4-[3-(TRIFLUOROMETHYL)-6,7-DIHYDROPYRANO[4,3-C]PYRAZOL-1(4H)-YL]PHENYL}METHYL)-2-PYRROLIDINONE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
1GME
 
 | | Crystal structure and assembly of an eukaryotic small heat shock protein | | Descriptor: | HEAT SHOCK PROTEIN 16.9B | | Authors: | Van Montfort, R.L.M, Basha, E, Friedrich, K.L, Slingsby, C, Vierling, E. | | Deposit date: | 2001-09-13 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Assembly of an Eukaryotic Small Heat Shock Protein
Nat.Struct.Biol., 8, 2001
|
|
1PKD
 
 | |
3R8I
 
 | | Crystal Structure of PPARgamma with an achiral ureidofibrate derivative (RT86) | | Descriptor: | 2-(4-{2-[1,3-benzoxazol-2-yl(heptyl)amino]ethyl}phenoxy)-2-methylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Laghezza, A, Fracchiolla, G, Lavecchia, A, Novellino, E, Crestani, M. | | Deposit date: | 2011-03-24 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, characterization and biological evaluation of ureidofibrate-like derivatives endowed with peroxisome proliferator-activated receptor activity.
J.Med.Chem., 55, 2012
|
|
3RWM
 
 | | Crystal Structure of Ypt32 in Complex with GppNHp | | Descriptor: | GTP-binding protein YPT32/YPT11, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Sultana, A, Dregger, C, Jin, Y, Franklin, E, Weisman, L.S, Khan, A.R. | | Deposit date: | 2011-05-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The activation cycle of Rab GTPase Ypt32 reveals structural determinants of effector recruitment and GDI binding.
Febs Lett., 585, 2011
|
|
3FFX
 
 | | Crystal Structure of CheY triple mutant F14E, N59R, E89H complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
1PAG
 
 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | FORMYCIN-5'-MONOPHOSPHATE, POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
3FGZ
 
 | | Crystal Structure of CheY triple mutant F14E, N59M, E89R complexed with BeF3- and Mn2+ | | Descriptor: | AMMONIUM ION, BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, ... | | Authors: | Wollish, A.C, Miller, P.J, Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
1QR1
 
 | | POOR BINDING OF A HER-2/NEU EPITOPE (GP2) TO HLA-A2.1 IS DUE TO A LACK OF INTERACTIONS IN THE CENTER OF THE PEPTIDE | | Descriptor: | BETA-2 MICROGLOBULIN, GP2 PEPTIDE, HLA-A2.1 HEAVY CHAIN | | Authors: | Kuhns, J.J, Batalia, M.A, Yan, S, Collins, E.J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-01-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide.
J.Biol.Chem., 274, 1999
|
|
1BZ9
 
 | |
2AAI
 
 | | Crystallographic refinement of ricin to 2.5 Angstroms | | Descriptor: | RICIN (A CHAIN), RICIN (B CHAIN), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rutenber, E, Katzin, B.J, Montfort, W, Villafranca, J.E, Ernst, S.R, Collins, E.J, Mlsna, D, Monzingo, A.F, Ready, M.P, Robertus, J.D. | | Deposit date: | 1993-09-07 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic refinement of ricin to 2.5 A.
Proteins, 10, 1991
|
|
1P5E
 
 | | The structure of phospho-CDK2/cyclin A in complex with the inhibitor 4,5,6,7-tetrabromobenzotriazole (TBS) | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, Cell division protein kinase 2, Cyclin A2 | | Authors: | De Moliner, E, Brown, N.R, Johnson, L.N. | | Deposit date: | 2003-04-26 | | Release date: | 2003-07-01 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Alternative binding modes of an inhibitor to two different kinases
Eur.J.Biochem., 270, 2003
|
|
1B0G
 
 | |
1DS5
 
 | | DIMERIC CRYSTAL STRUCTURE OF THE ALPHA SUBUNIT IN COMPLEX WITH TWO BETA PEPTIDES MIMICKING THE ARCHITECTURE OF THE TETRAMERIC PROTEIN KINASE CK2 HOLOENZYME. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CASEIN KINASE, ALPHA CHAIN, ... | | Authors: | Battistutta, R, Sarno, S, De Moliner, E, Marin, O, Zanotti, G, Pinna, L.A. | | Deposit date: | 2000-01-07 | | Release date: | 2001-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | The crystal structure of the complex of Zea mays alpha subunit with a fragment of human beta subunit provides the clue to the architecture of protein kinase CK2 holoenzyme.
Eur.J.Biochem., 267, 2000
|
|
