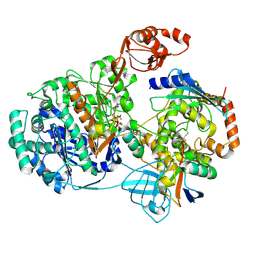
9B5D
 
 | |
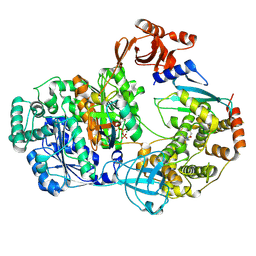
9B5C
 
 | |
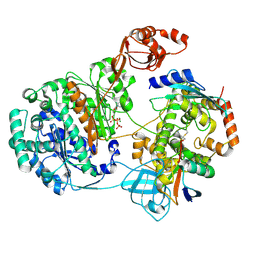
9B5N
 
 | |
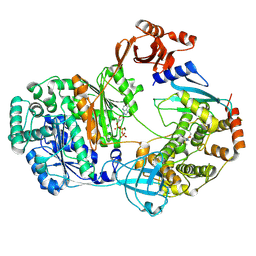
9B5R
 
 | |
9B5J
 
 | |
9B5U
 
 | |
9B5L
 
 | |
9B5A
 
 | |
9B5F
 
 | |
9B5H
 
 | |
9B5S
 
 | |
9B56
 
 | |
9B5I
 
 | |
9B5Q
 
 | |
9B5X
 
 | |
9B5B
 
 | |
9B5M
 
 | |
1TQ8
 
 | | Crystal Structure of protein Rv1636 from Mycobacterium tuberculosis H37Rv | | Descriptor: | hypothetical protein Rv1636 | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-16 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hypothetical protein Rv1636 from Mycobacterium tuberculosis H37Rv
To be Published
|
|
1TK9
 
 | | Crystal Structure of Phosphoheptose isomerase 1 | | Descriptor: | Phosphoheptose isomerase 1 | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two putative phosphoheptose isomerases.
Proteins, 63, 2006
|
|
1T35
 
 | | CRYSTAL STRUCTURE OF A HYPOTHETICAL PROTEIN YVDD- A PUTATIVE LYSINE DECARBOXYLASE | | Descriptor: | HYPOTHETICAL PROTEIN YVDD, Putative Lysine Decarboxylase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structure of a Hypothetical Protein Yvdd - Putative Lysine Decarboxylase
To be Published
|
|
1RI1
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI5
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI2
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
1RI4
 
 | | Structure and mechanism of mRNA cap (guanine N-7) methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, mRNA CAPPING ENZYME | | Authors: | Fabrega, C, Hausmann, S, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-11-16 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of mRNA cap (Guanine-n7) methyltransferase
Mol.Cell, 13, 2004
|
|
9B5V
 
 | |
