4B3G
 
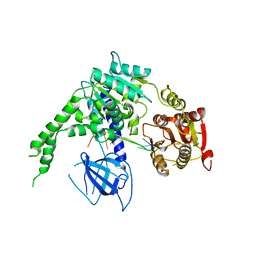 | | crystal structure of Ighmbp2 helicase in complex with RNA | | Descriptor: | DNA-BINDING PROTEIN SMUBP-2, RNA (5'-(AP*AP*AP*AP*AP*AP*AP*AP*AP)-3') | | Authors: | Lim, S.C, Song, H. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
4B3F
 
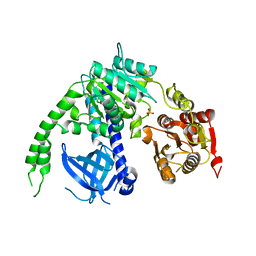 | | crystal structure of Ighmbp2 helicase | | Descriptor: | DNA-BINDING PROTEIN SMUBP-2, PHOSPHATE ION | | Authors: | Lim, S.C, Song, H. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1.
Nucleic Acids Res., 40, 2012
|
|
3M6N
 
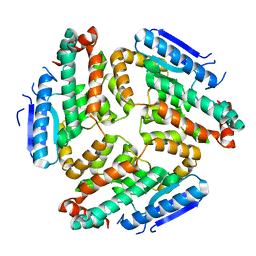 | | Crystal structure of RpfF | | Descriptor: | RpfF protein | | Authors: | Lim, S.C, Cheng, Z, Qamra, R, Song, H. | | Deposit date: | 2010-03-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Sensor-Synthase Interaction in Autoinduction of the Quorum Sensing Signal DSF Biosynthesis
Structure, 18, 2010
|
|
3TTN
 
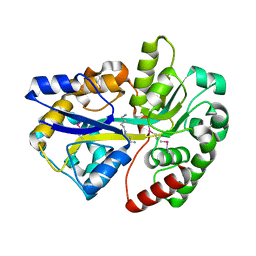 | |
3M6M
 
 | | Crystal structure of RpfF complexed with REC domain of RpfC | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Cheng, Z, Lim, S.C, Qamra, R, Song, H. | | Deposit date: | 2010-03-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Sensor-Synthase Interaction in Autoinduction of the Quorum Sensing Signal DSF Biosynthesis
Structure, 18, 2010
|
|
3TTM
 
 | | Crystal structure of SpuD in complex with putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
3TTL
 
 | | Crystal structure of apo-SpuE | | Descriptor: | Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
3TTK
 
 | | Crystal structure of apo-SpuD | | Descriptor: | Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
