1T9I
 
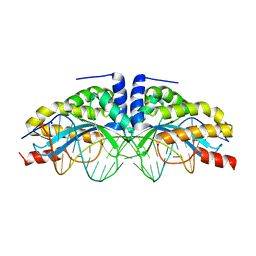 | | I-CreI(D20N)/DNA complex | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Sussman, D, Otis, C, Boudreau, D, Turmel, M, Lemieux, C, Stephens, K, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Dependent DNA Cleavage Mechanism of the I-CreI LAGLIDADG Homing Endonuclease.
Biochemistry, 43, 2004
|
|
1T9J
 
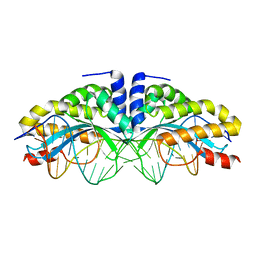 | | I-CreI(Q47E)/DNA complex | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', DNA endonuclease I-CreI | | Authors: | Chevalier, B, Sussman, D, Otis, C, Boudreau, D, Turmel, M, Lemieux, C, Stephens, K, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-Dependent DNA Cleavage Mechanism of the I-CreI LAGLIDADG Homing Endonuclease.
Biochemistry, 43, 2004
|
|
1MOW
 
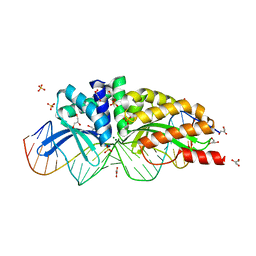 | | E-DreI | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*G)-3', 5'-D(*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', GLYCEROL, ... | | Authors: | Chevalier, B.S, Kortemme, T, Chadsey, M.S, Baker, D, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2002-09-10 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Activity and Structure of a Highly Specific Artificial Endonuclease
Mol.Cell, 10, 2002
|
|
1M5X
 
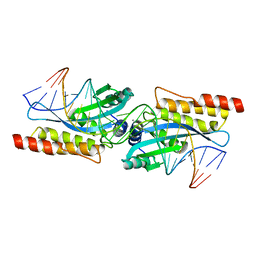 | | Crystal structure of the homing endonuclease I-MsoI bound to its DNA substrate | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-07-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
1N3E
 
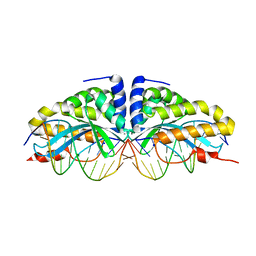 | | Crystal structure of I-CreI bound to a palindromic DNA sequence I (palindrome of left side of wildtype DNA target sequence) | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*C)-3', 5'-D(P*GP*AP*CP*GP*TP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
1N3F
 
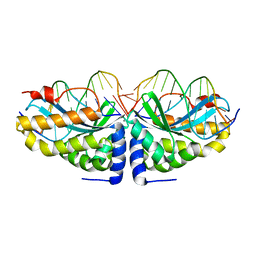 | | Crystal structure of I-CreI bound to a palindromic DNA sequence II (palindrome of right side of wildtype DNA target sequence) | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*GP*A)-3', 5'-D(P*GP*AP*CP*AP*GP*TP*TP*TP*CP*G-3'), CALCIUM ION, ... | | Authors: | Chevalier, B, Turmel, M, Lemieux, C, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexible DNA Target Site Recognition by Divergent Homing Endonuclease Isoschizomers I-CreI and I-MsoI
J.Mol.Biol., 329, 2003
|
|
1G9Y
 
 | | HOMING ENDONUCLEASE I-CREI / DNA SUBSTRATE COMPLEX WITH CALCIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', CALCIUM ION, ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
1G9Z
 
 | | LAGLIDADG HOMING ENDONUCLEASE I-CREI / DNA PRODUCT COMPLEX WITH MAGNESIUM | | Descriptor: | 5'-D(*CP*GP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*C)-3', 5'-D(*GP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*GP*A)-3', 5'-D(P*GP*AP*CP*AP*GP*TP*TP*TP*CP*G)-3', ... | | Authors: | Chevalier, B, Monnat, R.J, Stoddard, B.L. | | Deposit date: | 2000-11-28 | | Release date: | 2001-04-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites.
Nat.Struct.Biol., 8, 2001
|
|
1QAV
 
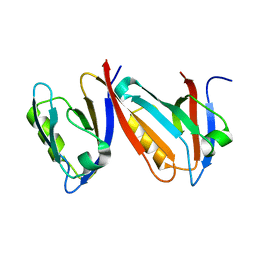 | | Unexpected Modes of PDZ Domain Scaffolding Revealed by Structure of NNOS-Syntrophin Complex | | Descriptor: | ALPHA-1 SYNTROPHIN (RESIDUES 77-171), NEURONAL NITRIC OXIDE SYNTHASE (RESIDUES 1-130) | | Authors: | Hillier, B.J, Christopherson, K.S, Prehoda, K.E, Bredt, D.S, Lim, W.A. | | Deposit date: | 1999-03-30 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex.
Science, 284, 1999
|
|
1QAU
 
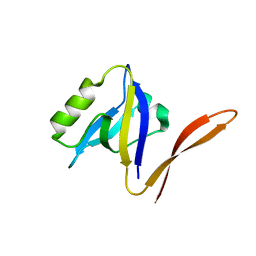 | | UNEXPECTED MODES OF PDZ DOMAIN SCAFFOLDING REVEALED BY STRUCTURE OF NNOS-SYNTROPHIN COMPLEX | | Descriptor: | NEURONAL NITRIC OXIDE SYNTHASE (RESIDUES 1-130) | | Authors: | Hillier, B.J, Christopherson, K.S, Prehoda, K.E, Bredt, D.S, Lim, W.A. | | Deposit date: | 1999-03-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex.
Science, 284, 1999
|
|
6XUC
 
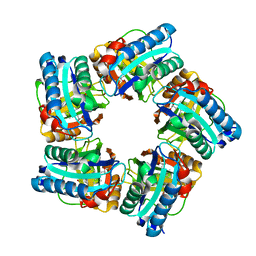 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with coproheme | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Chlorite dismutase | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8702 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
6XUB
 
 | | Structure of coproheme decarboxylase from Corynebacterium diphteriae in complex with monovinyl monopropionyl deuteroheme | | Descriptor: | Chlorite dismutase, harderoheme (III) | | Authors: | Michlits, H, Lier, B, Pfanzagl, V, Djinovic-Carugo, K, Furtmueller, P.G, Oostenbrink, C, Obinger, C, Hofbauer, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Actinobacterial Coproheme Decarboxylases Use Histidine as a Distal Base to Promote Compound I Formation.
Acs Catalysis, 10, 2020
|
|
1EVW
 
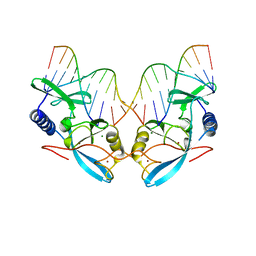 | | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*A)-3'), DNA (5'-D(*TP*GP*GP*CP*TP*AP*CP*CP*TP*TP*AP*A)-3'), DNA (5'-D(P*GP*AP*GP*AP*GP*TP*CP*A)-3'), ... | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
1EVX
 
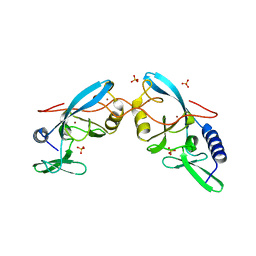 | | APO CRYSTAL STRUCTURE OF THE HOMING ENDONUCLEASE, I-PPOI | | Descriptor: | INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SULFATE ION, ZINC ION | | Authors: | Galburt, E.A, Jurica, M.S, Chevalier, B.S, Erho, D, Stoddard, B.L. | | Deposit date: | 2000-04-20 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site.
J.Mol.Biol., 300, 2000
|
|
6SHR
 
 | | X-RAY CRYSTAL STRUCTURE OF CELL-FREE PROTEIN SYNTHESIS (CFPS) PRODUCED SDF1-A | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Jugnarain, V.M, Mitchell, E, Forsyth, T, Michael, H, Cortes, S, Tillier, B. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | X-RAY CRYSTAL STRUCTURE OF CELL-FREE PROTEIN SYNTHESIS (CFPS) PRODUCED SDF1-A
To Be Published
|
|
1CYQ
 
 | | INTRON ENCODED HOMING ENDONUCLEASE I-PPOI (H98A)/DNA HOMING SITE COMPLEX | | Descriptor: | 5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3', INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, MAGNESIUM ION, ... | | Authors: | Galburt, E.A, Chevalier, B, Jurica, M.S, Flick, K.E, Stoddard, B.L. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A novel endonuclease mechanism directly visualized for I-PpoI.
Nat.Struct.Biol., 6, 1999
|
|
1CZ0
 
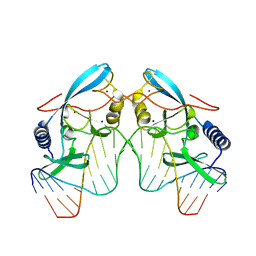 | | INTRON ENCODED HOMING ENDONUCLEASE I-PPOI/DNA COMPLEX LACKING CATALYTIC METAL ION | | Descriptor: | DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED HOMING ENDONUCLEASE I-PPOI, SODIUM ION, ... | | Authors: | Galburt, E.A, Chevalier, B, Jurica, M.S, Flick, K.E, Stoddard, B.L. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel endonuclease mechanism directly visualized for I-PpoI.
Nat.Struct.Biol., 6, 1999
|
|
4MLL
 
 | | The 1.4 A structure of the class D beta-lactamase OXA-1 K70D complexed with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase OXA-1, ... | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2013-09-06 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural Origins of Oxacillinase Specificity in Class D beta-Lactamases.
Antimicrob.Agents Chemother., 58, 2014
|
|
2MG6
 
 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [3,16]-trans dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
2MFX
 
 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-cis dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
2MFY
 
 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-trans dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
4F94
 
 | | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Beta-lactamase, SULFATE ION | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2012-05-18 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin
To be Published
|
|
