1V6N
 
 | | Peanut lectin with 9mer peptide (PVIWSSATG) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SFY
 
 | | Crystal structure of recombinant Erythrina corallodandron Lectin | | Descriptor: | CALCIUM ION, Lectin, MANGANESE (II) ION, ... | | Authors: | Kulkarni, K.A, Srivastava, A, Mitra, N, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2004-02-21 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effect of glycosylation on the structure of Erythrina corallodendron lectin.
Proteins, 56, 2004
|
|
1V6M
 
 | | Peanut Lectin with 9mer peptide (IWSSAGNVA) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UMN
 
 | | Crystal structure of Dps-like peroxide resistance protein (Dpr) from Streptococcus suis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kauko, A, Haataja, S, Pulliainen, A, Finne, J, Papageorgiou, A.C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation.
J. Mol. Biol., 338, 2004
|
|
2O4N
 
 | | Crystal Structure of HIV-1 Protease (TRM Mutant) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
2O4S
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Lopinavir | | Descriptor: | CHLORIDE ION, GLYCEROL, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
2M5R
 
 | |
1YRK
 
 | | The C2 Domain of PKC is a new Phospho-Tyrosine Binding Domain | | Descriptor: | 13-residue peptide, ACETIC ACID, Protein kinase C, ... | | Authors: | Benes, C.H, Wu, N, Elia, A.E, Dharia, T, Cantley, L.C, Soltoff, S.P. | | Deposit date: | 2005-02-03 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C2 domain of PKCdelta is a phosphotyrosine binding domain.
Cell(Cambridge,Mass.), 121, 2005
|
|
2NSD
 
 | |
7V4Z
 
 | | Structure of Horcolin native form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bobbili, K.B, Sivaji, N, Jayaprakash, N.G, Narayanan, V, Sekhar, A, Suguna, K, Surolia, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure and Carbohydrate Recognition by the Nonmitogenic Lectin Horcolin.
Biochemistry, 61, 2022
|
|
4PN7
 
 | |
4R6Q
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6R
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7V4S
 
 | | Horcolin complex with methyl-alpha-mannose | | Descriptor: | Horcolin, methyl alpha-D-mannopyranoside | | Authors: | Bobbili, K.B, Sivaji, N, Jayaprakash, N.G, Narayanan, V, Sekhar, A, Suguna, K, Surolia, A. | | Deposit date: | 2021-08-14 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and Carbohydrate Recognition by the Nonmitogenic Lectin Horcolin.
Biochemistry, 61, 2022
|
|
4P2E
 
 | | Acoustic transfer of protein crystals from agar pedestals to micromeshes for high throughput screening of heavy atom derivatives | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme C, ... | | Authors: | Cuttitta, C.M, Roessler, C.G, Ericson, D.L, Scalia, A, Teplitsky, E, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-03-03 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acoustic transfer of protein crystals from agarose pedestals to micromeshes for high-throughput screening.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4P9D
 
 | |
4OKC
 
 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
5OVN
 
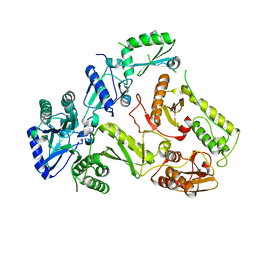 | | Crystal Structure of FIV Reverse Transcriptase | | Descriptor: | POL protein | | Authors: | Galilee, M, Alian, A. | | Deposit date: | 2017-08-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.942 Å) | | Cite: | The structure of FIV reverse transcriptase and its implications for non-nucleoside inhibitor resistance.
PLoS Pathog., 14, 2018
|
|
4LTE
 
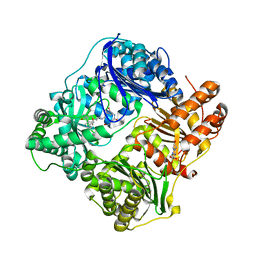 | | Structure of Cysteine-free Human Insulin Degrading Enzyme in Complex with Macrocyclic Inhibitor | | Descriptor: | 2,6-DIAMINO-HEXANOIC ACID AMIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FUMARIC ACID, ... | | Authors: | Foda, Z.H, Seeliger, M.A, Saghatelian, A, Liu, D.R. | | Deposit date: | 2013-07-23 | | Release date: | 2014-05-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones.
Nature, 511, 2014
|
|
4M65
 
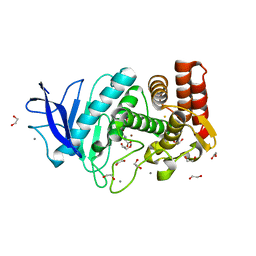 | | In situ thermolysin crystallized on a MiTeGen micromesh with asparagine ligand | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ
co-crystallization of proteins plus fragment libraries on
pin-mounted data-collection micromeshes
Acta Crystallogr.,Sect.D, D70
|
|
4P9C
 
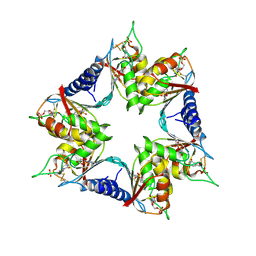 | |
1CQ9
 
 | | PEANUT LECTIN-TRICLINIC FORM | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN) | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-06 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual
quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry
and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
