8X5D
 
 | |
5ZUM
 
 | | Structure of dipeptidyl-peptidase III from Corallococcus sp. strain EGB | | 分子名称: | ZINC ION, dipeptidyl-peptidase III | | 著者 | Zhang, H, Duan, Y.J, Li, Z.K, Liu, W.D, Huang, Y, Cui, Z.L. | | 登録日 | 2018-05-08 | | 公開日 | 2019-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of dipeptidyl peptidase III from Corallococcus sp. strain EGB
To Be Published
|
|
6AIN
 
 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, PnpA | | 著者 | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | 登録日 | 2018-08-24 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6AIO
 
 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | 分子名称: | PnpA | | 著者 | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | 登録日 | 2018-08-24 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
7XE0
 
 | | Cryo-EM structure of plant NLR Sr35 resistosome | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, Sr35 | | 著者 | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | 登録日 | 2022-03-29 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7XVG
 
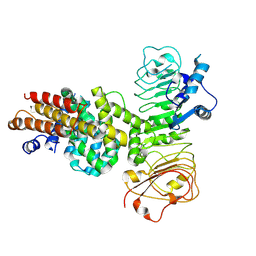 | | Cryo-EM structure of binary complex of plant NLR Sr35 and effector AvrSr35 | | 分子名称: | AvrSr35, Sr35 | | 著者 | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | 登録日 | 2022-05-23 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7XX2
 
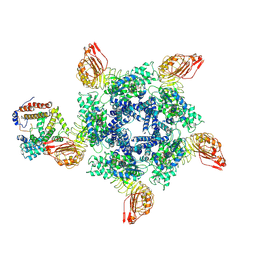 | | Cryo-EM structure of Sr35 resistosome induced by AvrSr35 R381A | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, AvrSr35, CNL9 | | 著者 | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | 登録日 | 2022-05-28 | | 公開日 | 2022-11-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
8I3Q
 
 | |
