8K4A
 
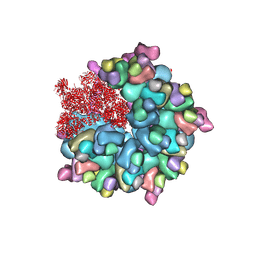 | | Structure of Banna virus core | | Descriptor: | VP10, VP2, VP8 | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8K44
 
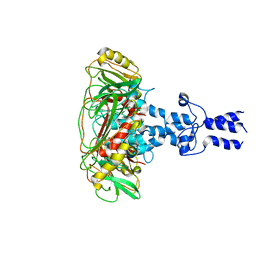 | | Structure of VP9 in Banna virus | | Descriptor: | VP9 | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8HBN
 
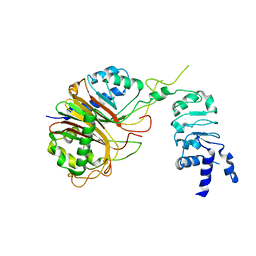 | | Structure of the Mex67-Mtr2-1 heterodimer | | Descriptor: | mRNA export factor MEX67, mRNA transport regulator MTR2 | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-10-29 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
7EJV
 
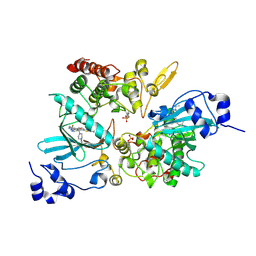 | | The co-crystal structure of DYRK2 with YK-2-69 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [6-[[4-[2-(dimethylamino)-1,3-benzothiazol-6-yl]-5-fluoranyl-pyrimidin-2-yl]amino]pyridin-3-yl]-(4-ethylpiperazin-1-yl)methanone | | Authors: | Li, Z, Xiao, Y, Yuan, K, Kuang, W, Xiuquan, Y, Yang, P. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting dual-specificity tyrosine phosphorylation-regulated kinase 2 with a highly selective inhibitor for the treatment of prostate cancer.
Nat Commun, 13, 2022
|
|
5JJO
 
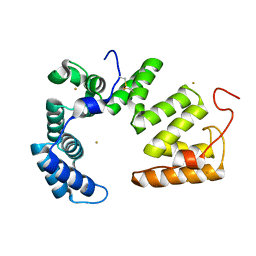 | | The crystal structure of immunity protein PA5088 from Pseudomonas aeruginosa | | Descriptor: | GOLD ION, Uncharacterized protein | | Authors: | Li, Z.Q, Gao, Z.Q, She, Z, Dong, Y.H. | | Deposit date: | 2016-04-24 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural analysis of Pseudomonas aeruginosa H3-T6SS immunity proteins
Febs Lett., 590, 2016
|
|
5JKP
 
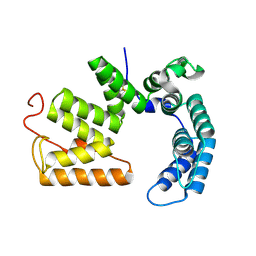 | |
5XJ5
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the monoacylglycerol form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLYCINE, Glycerol-3-phosphate acyltransferase, ... | | Authors: | Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
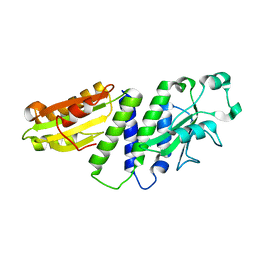
4ETX
 
 | |
4EUV
 
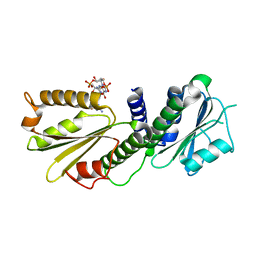 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1, in complex with c-di-GMP, form 1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-25 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
8K43
 
 | |
4EU0
 
 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
4ETZ
 
 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
7Y5M
 
 | |
4J8C
 
 | |
8HPD
 
 | | Bry-LHCII heterotrimer of Bryopsis corticulans | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHLOROPHYLL A, ... | | Authors: | Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional properties of different types of siphonous LHCII trimers from an intertidal green alga Bryopsis corticulans.
Structure, 31, 2023
|
|
4J8E
 
 | |
4J8D
 
 | |
6AG0
 
 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | Authors: | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
7X7W
 
 | | The X-ray Crystallographic Structure of D-Psicose 3-epimerase from Clostridia bacterium | | Descriptor: | D-PSICOSE 3-EPIMERASE, MANGANESE (II) ION | | Authors: | Li, Z.F, Ban, X.F, Xie, X.F, Tian, Y.X, Li, C.M, Gu, Z.B. | | Deposit date: | 2022-03-10 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of a novel homodimeric D-allulose 3-epimerase from a Clostridia bacterium
Acta Crystallogr.,Sect.D, 78, 2022
|
|
8K42
 
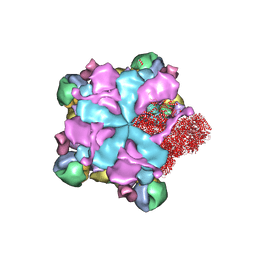 | | Structure of full Banna virus | | Descriptor: | VP10, VP2, VP4, ... | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
6J3X
 
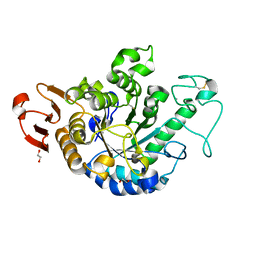 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotriose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-01-06 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
7VMA
 
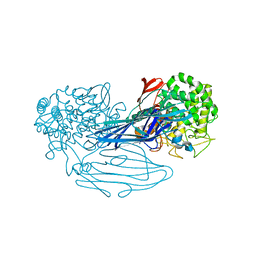 | | The X-ray crystallographic structure of amylo-alpha-1,6-glucosidase from Thermococcus gammatolerans STB12 | | Descriptor: | Amylo-alpha-1,6-glucosidase, putative archaeal type glycogen debranching enzyme (Gde) | | Authors: | Li, Z.F, Ban, X.F, Wang, Y.M, Li, C.M, Gu, Z.B. | | Deposit date: | 2021-10-08 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | The X-ray Crystallographic Structure of Amylo-alpha-1,6-glucosidase from Thermococcus gannatilerans STB12
To Be Published
|
|
6IGC
 
 | | Crystal structure of HPV58/33/52 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IYG
 
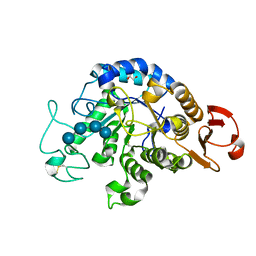 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-15 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
