6NYF
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 1 (OA-1) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1H52
 
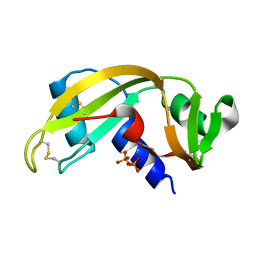 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PYROPHOSPHATE 2- | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
5WSN
 
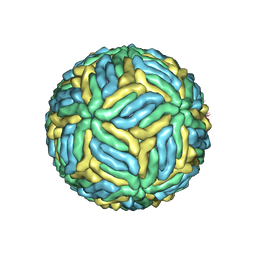 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
6NYM
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2d (OA-2d) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYJ
 
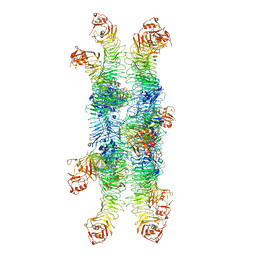 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2b (OA-2b) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1WM3
 
 | |
1H53
 
 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, CITRIC ACID, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1FNJ
 
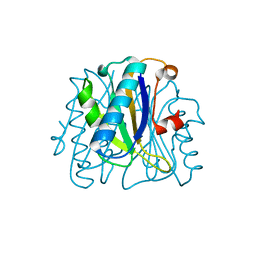 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88S/R90K | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
5D1M
 
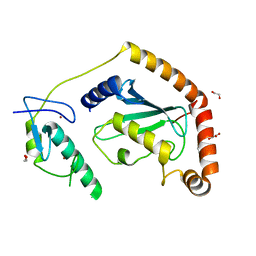 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 (P199A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
1WM2
 
 | |
3UIU
 
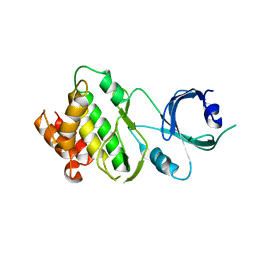 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
7K3V
 
 | | Apoferritin structure at 1.34 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with K3 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.34 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
1Q34
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2-21.5 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
7K3W
 
 | | Apoferritin structure at 1.36 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with Falcon4 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.36 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
1FNK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88K/R90S | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1OOJ
 
 | | Structural genomics of Caenorhabditis elegans : Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin CMD-1 | | Authors: | Symersky, J, Lin, G, Li, S, Qiu, S, Luan, C.-H, Luo, D, Tsao, J, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural genomics of caenorhabditis elegans: crystal structure of calmodulin.
Proteins, 53, 2003
|
|
6VQV
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQX
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7PVN
 
 | | Crystal Structure of Human UBA6 in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Truongvan, N, Li, S, Schindelin, H. | | Deposit date: | 2021-10-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
4WVR
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
7R7S
 
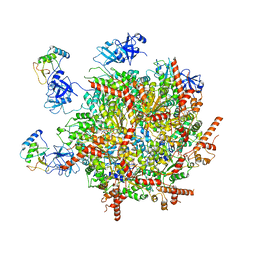 | | p47-bound p97-R155H mutant with ATPgammaS | | Descriptor: | NSFL1 cofactor p47, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7T
 
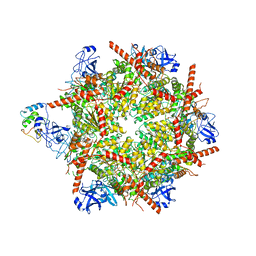 | | p47-bound p97-R155H mutant with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NSFL1 cofactor p47, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7R7U
 
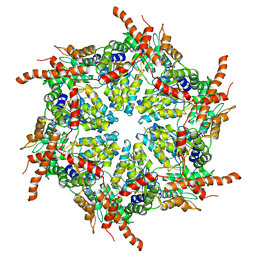 | | D1 and D2 domain structure of the p97(R155H)-p47 complex | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2021-06-25 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
