5FTE
 
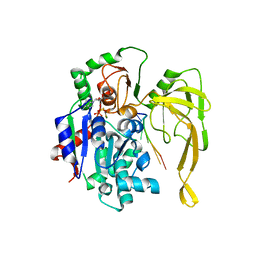 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP-AlF3 and ssDNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
2KDH
 
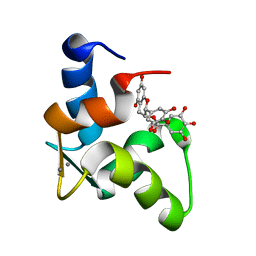 | | The Solution Structure of Human Cardiac Troponin C in complex with the Green Tea Polyphenol; (-)-epigallocatechin-3-gallate | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, CALCIUM ION, Troponin C, ... | | Authors: | Robertson, I.M, Li, M.X, Sykes, B.D. | | Deposit date: | 2009-01-09 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human cardiac troponin C in complex with the green tea polyphenol, (-)-epigallocatechin 3-gallate
J.Biol.Chem., 284, 2009
|
|
1JL9
 
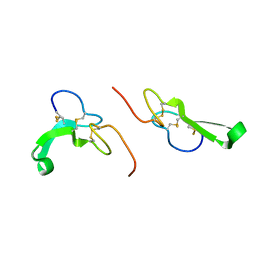 | | Crystal Structure of Human Epidermal Growth Factor | | Descriptor: | EPIDERMAL GROWTH FACTOR | | Authors: | Lu, H.S, Chai, J.J, Li, M, Huang, B.R, He, C.H, Bi, R.C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human epidermal growth factor and its dimerization
J.Biol.Chem., 276, 2001
|
|
5FTC
 
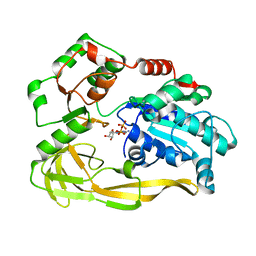 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTD
 
 | | Crystal structure of Pif1 helicase from Bacteroides apo form | | Descriptor: | PHOSPHATE ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTF
 
 | | Crystal structure of Pif1 helicase from Bacteroides double mutant L95C-I339C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
4RTH
 
 | | The crystal structure of PsbP from Zea mays | | Descriptor: | Membrane-extrinsic protein of photosystem II PsbP | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
4RT4
 
 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
4RTI
 
 | | The crystal structure of PsbP from Spinacia oleracea | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Oxygen-evolving enhancer protein 2, ... | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
4RTA
 
 | |
1B8Q
 
 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
1LVM
 
 | | CATALYTICALLY ACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH PRODUCT | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
8X7T
 
 | | MCM in the Apo state. | | Descriptor: | mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | MCM in the Apo state
To Be Published
|
|
5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
1YGJ
 
 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | (2R)-2-({6-[BENZYL(METHYL)AMINO]-9-ISOPROPYL-9H-PURIN-2-YL}AMINO)BUTAN-1-OL, Pyridoxal kinase | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives
J.Biol.Chem., 280, 2005
|
|
1YGK
 
 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | Pyridoxal kinase, R-ROSCOVITINE | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives
J.Biol.Chem., 280, 2005
|
|
1YHJ
 
 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | (2R)-2-{[6-(BENZYLOXY)-9-ISOPROPYL-9H-PURIN-2-YL]AMINO}BUTAN-1-OL, Pyridoxal Kinase | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-09 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives.
J.Biol.Chem., 280, 2005
|
|
4YZH
 
 | | Structure of the Arabidopsis TAP38/PPH1 in complex with pLhcb1 phosphopeptide substrate | | Descriptor: | Chlorophyll a-b binding protein 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Wei, X.P, Guo, J.T, Li, M, Liu, Z.F. | | Deposit date: | 2015-03-25 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Mechanism Underlying the Specific Recognition between the Arabidopsis State-Transition Phosphatase TAP38/PPH1 and Phosphorylated Light-Harvesting Complex Protein Lhcb1
Plant Cell, 27, 2015
|
|
2L1R
 
 | | The structure of the calcium-sensitizer, dfbp-o, in complex with the N-domain of troponin C and the switch region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Robertson, I.M, Sun, Y, Li, M.X, Sykes, B.D. | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural and functional perspective into the mechanism of Ca(2+)-sensitizers that target the cardiac troponin complex.
J.MOL.CELL.CARDIOL., 49, 2010
|
|
5XNL
 
 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
3K0T
 
 | |
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
1PVZ
 
 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
1LVB
 
 | | CATALYTICALLY INACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH SUBSTRATE | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), GLYCEROL, OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
