6KQE
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 4 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQM
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 5 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQG
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 6 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQL
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 4 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5XYZ
 
 | | The structure of human BTK kinase domain in complex with a covalent inhibitor | | Descriptor: | N-[3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-1H-1,2,4-triazol-3-yl)phenyl]propanamide, Tyrosine-protein kinase BTK | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Xie, Y.T, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
6KQH
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 7 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQN
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 6 nt | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.489 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQF
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 5 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6J1K
 
 | | Crystal structure of ribose-5-phosphate isomerase A from Ochrobactrum sp. CSL1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ribose-5-phosphate isomerase A | | Authors: | Xu, X.Q, Mo, X.B, Ju, X, Shen, M, Li, L.Z, Yuan, A.Y. | | Deposit date: | 2018-12-28 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | Crystal structure of ribose-5-phosphate isomerase A from Ochrobactrum sp. CSL1
To be published
|
|
6JUY
 
 | | Crystal Structure of ArgZ, apo structure, an Arginine Dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JV1
 
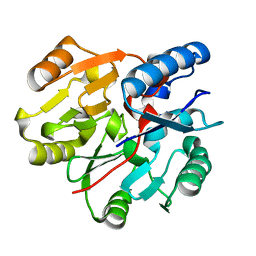 | | Crystal Structure of N-terminal domain of ArgZ, C264S mutant, bound to Substrate, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
7DCD
 
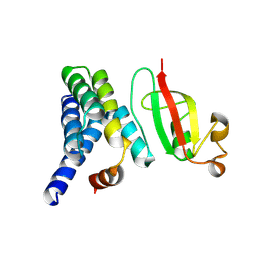 | |
6LP3
 
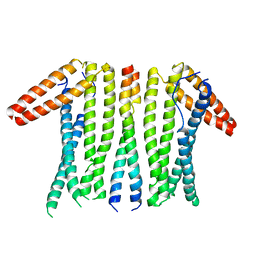 | | Structural basis and functional analysis epo1-bem3p complex for bud growth | | Descriptor: | GTPase-activating protein BEM3, Uncharacterized protein YMR124W | | Authors: | Wang, J, Li, L, Ming, Z.H, Wu, L.J, Yan, L.M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Structural basis and functional analysis epo1-bem3p complex for bud growth
To Be Published
|
|
7CMO
 
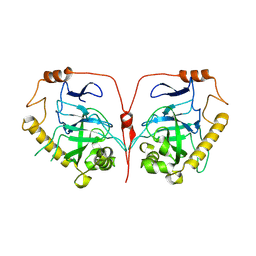 | | Crystal structure of human inorganic pyrophosphatase | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Hu, F, Huang, Z, Li, L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biochemical characterization of inorganic pyrophosphatase from Homo sapiens.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CP2
 
 | | Crystal structure of the African swine fever virus core shell protein p15 | | Descriptor: | CP530R | | Authors: | Liu, K.F, Meng, Y.M, Chai, Y, Li, L.J, Sun, H, Gao, G.F, Tan, S.G, Qi, J.X. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the African swine fever virus core shell protein p15
Biosaf Health, 2021
|
|
4PKM
 
 | | Crystal Structure of Bacillus thuringiensis Cry51Aa1 Protoxin at 1.65 Angstroms Resolution | | Descriptor: | Cry51Aa1, GLYCEROL, GLYCINE, ... | | Authors: | Xu, C, Chinte, U, Chen, L, Yao, Q, Zhou, D, Meng, Y, Li, L, Rose, J, Bi, L.J, Yu, Z, Sun, M, Wang, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Cry51Aa1: A potential novel insecticidal aerolysin-type beta-pore-forming toxin from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 462, 2015
|
|
6PH1
 
 | | T4 lysozyme pseudo-wild type soaked in TEMPOL | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6PGZ
 
 | | MTSL labelled T4 lysozyme pseudo-wild type V75C mutant | | Descriptor: | CHLORIDE ION, Endolysin, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6PH0
 
 | | T4 lysozyme pseudo-wild type soaked in TEMPO | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6PGY
 
 | |
7EGS
 
 | | The crystal structure of lobe domain of E. coli RNA polymerase complexed with the C-terminal domain of UvrD | | Descriptor: | DNA helicase II, DNA-directed RNA polymerase subunit beta, GLYCEROL | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
7EGT
 
 | | The crystal structure of the C-terminal domain of T. thermophilus UvrD complexed with the N-terminal domain of UvrB | | Descriptor: | DNA helicase UvrD, UvrABC system protein B | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
