1VRY
 
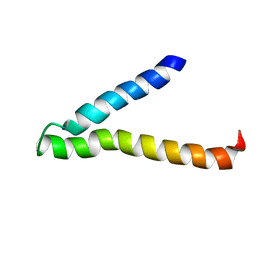 | | Second and Third Transmembrane Domains of the Alpha-1 Subunit of Human Glycine Receptor | | 分子名称: | Glycine receptor alpha-1 chain | | 著者 | Ma, D, Liu, Z, Li, L, Tang, P, Xu, Y. | | 登録日 | 2005-07-20 | | 公開日 | 2005-07-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and Dynamics of the Second and Third Transmembrane Domains of Human Glycine Receptor.
Biochemistry, 44, 2005
|
|
6U0F
 
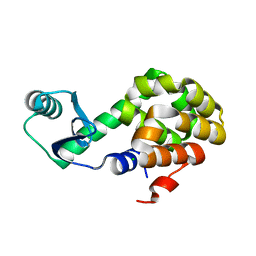 | |
6U0B
 
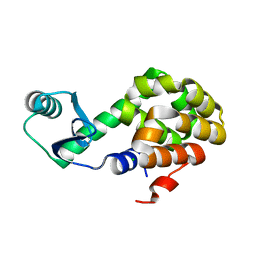 | |
6U0C
 
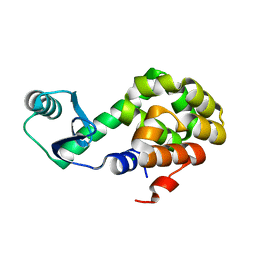 | |
3II9
 
 | | Crystal structure of glutaryl-coa dehydrogenase from Burkholderia pseudomallei at 1.73 Angstrom | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutaryl-CoA dehydrogenase, ... | | 著者 | Ismagilov, R.F, Li, L, Du, W.B, Staker, B, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2009-07-31 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments.
J.Am.Chem.Soc., 132, 2010
|
|
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | 分子名称: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | 著者 | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | 登録日 | 2017-09-15 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | 分子名称: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | 著者 | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | 登録日 | 2017-09-15 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6VRJ
 
 | |
2HVV
 
 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | 分子名称: | SULFATE ION, ZINC ION, deoxycytidylate deaminase | | 著者 | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | 登録日 | 2006-07-31 | | 公開日 | 2007-09-11 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
2HVW
 
 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ... | | 著者 | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | 登録日 | 2006-07-31 | | 公開日 | 2007-09-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
4FQB
 
 | |
3L8R
 
 | |
6LCR
 
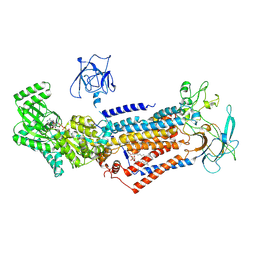 | | Cryo-EM structure of Dnf1 from Chaetomium thermophilum in the E1-ATP state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cdc50, ... | | 著者 | He, Y, Xu, J, Wu, X, Li, L. | | 登録日 | 2019-11-19 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of a P4-ATPase lipid flippase in lipid bilayers.
Protein Cell, 11, 2020
|
|
4OUL
 
 | |
5X8I
 
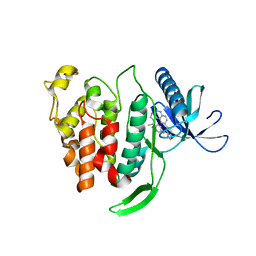 | | Crystal structure of human CLK1 in complex with compound 25 | | 分子名称: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | 著者 | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | 登録日 | 2017-03-02 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | 著者 | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | 登録日 | 2017-05-15 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
4IYR
 
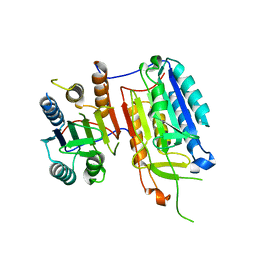 | | Crystal structure of full-length caspase-6 zymogen | | 分子名称: | Caspase-6 | | 著者 | Cao, Q, Wang, X.-J, Li, L.-F, Su, X.-D. | | 登録日 | 2013-01-29 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.697 Å) | | 主引用文献 | The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6JV0
 
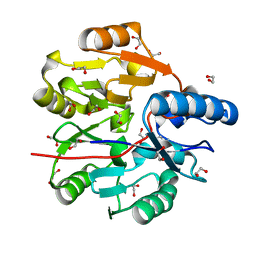 | | Crystal Structure of N-terminal domain of ArgZ, bound to Product, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | 分子名称: | 1,2-ETHANEDIOL, L-ornithine, Sll1336 protein | | 著者 | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | 登録日 | 2019-04-15 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
5ZIU
 
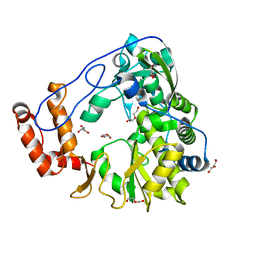 | | Crystal structure of human Entervirus D68 RdRp | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, RdRp | | 著者 | Wang, M.L, Zhang, Y, Chen, Y.P, Lu, D.R, Jiang, H, Chen, Y.J, Li, L, Zhang, C.H, Shi, Q.L, Su, D. | | 登録日 | 2018-03-17 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Crystal structure of human Entervirus D68 RdRp in complex with NADPH
To Be Published
|
|
6L74
 
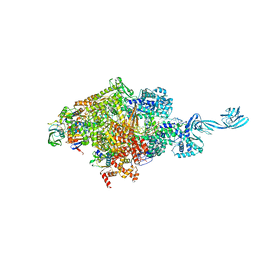 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 2 nt | | 分子名称: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Zhang, Y, Li, L, Ebright, R.H. | | 登録日 | 2019-10-31 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L4R
 
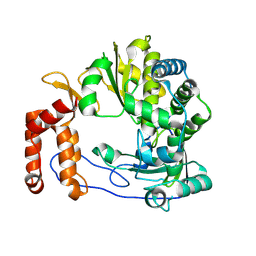 | | Crystal structure of Enterovirus D68 RdRp | | 分子名称: | RdRp | | 著者 | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | 登録日 | 2019-10-21 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
3F9G
 
 | |
3F9F
 
 | |
3F9H
 
 | |
3CXS
 
 | | Crystal structure of human GNA1 | | 分子名称: | Glucosamine 6-phosphate N-acetyltransferase | | 著者 | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | 登録日 | 2008-04-25 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
