4O7P
 
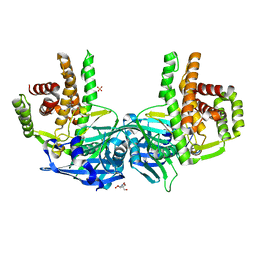 | | Crystal structure of Mycobacterium tuberculosis maltose kinase MaK complexed with maltose | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Maltokinase, SULFATE ION, ... | | Authors: | Li, J, Guan, X.T, Rao, Z.H. | | Deposit date: | 2013-12-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Homotypic dimerization of a maltose kinase for molecular scaffolding.
Sci Rep, 4, 2014
|
|
8QEW
 
 | |
6ABO
 
 | | human XRCC4 and IFFO1 complex | | Descriptor: | DNA repair protein XRCC4, GLYCEROL, Intermediate filament family orphan 1, ... | | Authors: | Li, J, Liu, L, Liang, H, Liu, Y, Xu, D. | | Deposit date: | 2018-07-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Nat.Cell Biol., 21, 2019
|
|
4O7O
 
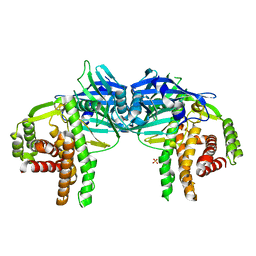 | |
3RY7
 
 | | Crystal Structure of Sa239 | | Descriptor: | GLYCEROL, Ribokinase | | Authors: | Li, J, Wu, M, Wang, L, Zang, J. | | Deposit date: | 2011-05-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Sa239 reveals the structural basis for the activation of ribokinase by monovalent cations.
J.Struct.Biol., 177, 2012
|
|
5YTK
 
 | | Crystal structure of SIRT3 bound to a leucylated AceCS2 | | Descriptor: | AceCS2-KLeu, LEUCINE, LYSINE, ... | | Authors: | Li, J, Gong, W, Xu, Y. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations
Cell Metab., 27, 2018
|
|
5HC3
 
 | | The structure of esterase Est22 | | Descriptor: | GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC2
 
 | |
4L93
 
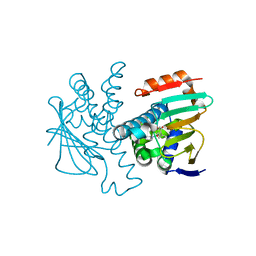 | | Crystal structure of Human Hsp90 with S36 | | Descriptor: | 3,4-dihydroisoquinolin-2(1H)-yl[2,4-dihydroxy-5-(propan-2-yl)phenyl]methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Crystal structure of Human Hsp90 with S36
To be Published
|
|
4L90
 
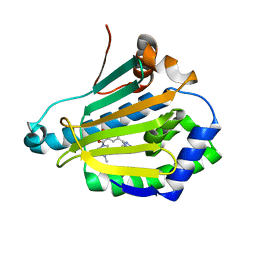 | | Crystal structure of Human Hsp90 with RL3 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](4-methylpiperazin-1-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Human Hsp90 with RL3
to be published
|
|
5HC5
 
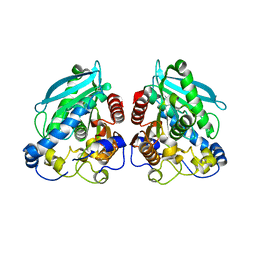 | |
5HC4
 
 | | Structure of esterase Est22 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC0
 
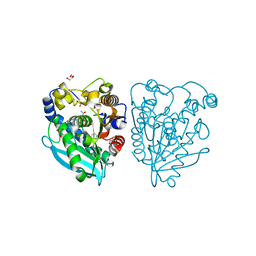 | | Structure of esterase Est22 with p-nitrophenol | | Descriptor: | ACETIC ACID, GLYCEROL, Lipolytic enzyme, ... | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
2ELA
 
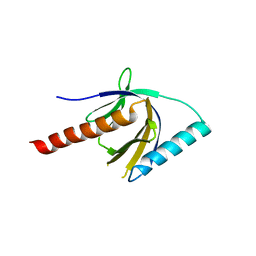 | | Crystal Structure of the PTB domain of human APPL1 | | Descriptor: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | Authors: | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
4LWE
 
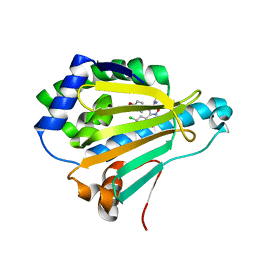 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-(5-chloro-2,4-dihydroxyphenyl)-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]acetamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWF
 
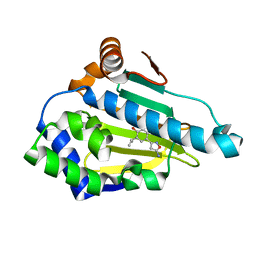 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ3 | | Descriptor: | 4-(5-amino-1,2-oxazol-3-yl)-6-(propan-2-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWI
 
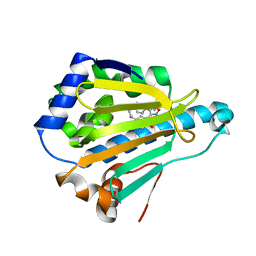 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
2ELB
 
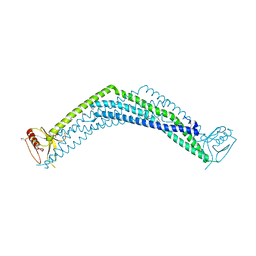 | | Crystal Structure of the BAR-PH domain of human APPL1 | | Descriptor: | Adapter protein containing PH domain, PTB domain and leucine zipper motif 1 | | Authors: | Li, J, Mao, X, Dong, L.Q, Liu, F, Tong, L. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the BAR-PH and PTB Domains of Human APPL1
Structure, 15, 2007
|
|
2B26
 
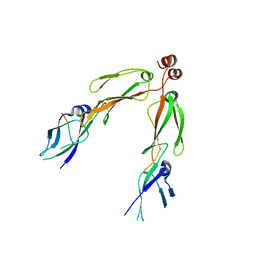 | | The crystal structure of the protein complex of yeast Hsp40 Sis1 and Hsp70 Ssa1 | | Descriptor: | Heat shock 70 kDa protein cognate 2, SIS1 protein | | Authors: | Li, J, Wu, Y, Qian, X, Sha, B. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex.
Biochem.J., 398, 2006
|
|
4LWG
 
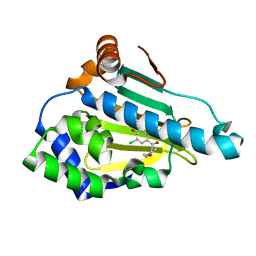 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ4 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-2-(4-methoxyphenyl)ethanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWH
 
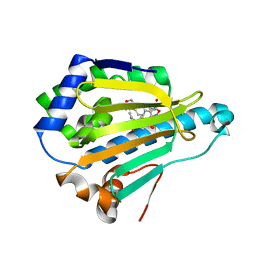 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
2FCO
 
 | | Crystal Structure of Bacillus stearothermophilus PrfA-Holliday Junction Resolvase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, recombination protein U (penicillin-binding protein related factor A) | | Authors: | Li, J, Jedrzejas, M.J. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, flexibility, and mechanism of the Bacillus stearothermophilus RecU Holliday junction resolvase.
Proteins, 68, 2007
|
|
6M3P
 
 | | Crystal structure of AnkG/beta2-spectrin complex | | Descriptor: | Ankyrin-3, Spectrin beta chain, non-erythrocytic 1 | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
8EW5
 
 | | The structure of flightin within myosin thick filaments from Bombus ignitus flight muscle | | Descriptor: | Flightin | | Authors: | Li, J, Rahmani, H, Abbasi Yeganeh, F, Rastegarpouyani, H, Taylor, D.W, Wood, N.B, Previs, M.J, Iwamoto, H, Taylor, K.A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus , at 6 angstrom Resolution.
Int J Mol Sci, 24, 2022
|
|
1SEK
 
 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | Descriptor: | SERPIN K | | Authors: | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
