7N17
 
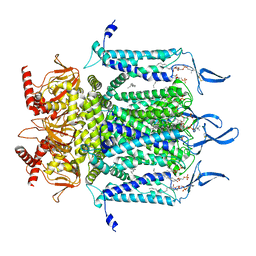 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
6DJV
 
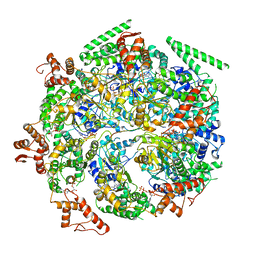 | | Mtb ClpB in complex with ATPgammaS and casein, Conformer 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2018-05-26 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ATP hydrolysis-coupled peptide translocation mechanism ofMycobacterium tuberculosisClpB.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8FS5
 
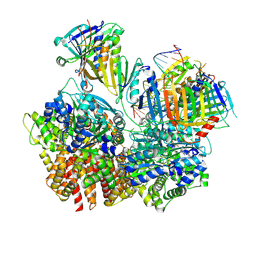 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS3
 
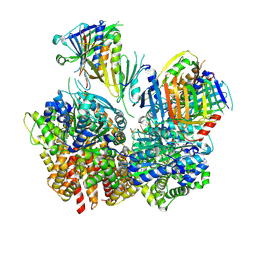 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS4
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS7
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS8
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS6
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
6DJU
 
 | | Mtb ClpB in complex with ATPgammaS and casein, Conformer 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2018-05-26 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ATP hydrolysis-coupled peptide translocation mechanism ofMycobacterium tuberculosisClpB.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4QBS
 
 | |
5I0A
 
 | | RecA mini intein in complex with cisplatin | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Cisplatin, Intein, ... | | Authors: | Li, Z, Zhang, J, Li, H.M. | | Deposit date: | 2016-02-03 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
2R41
 
 | |
4QBR
 
 | |
2R7A
 
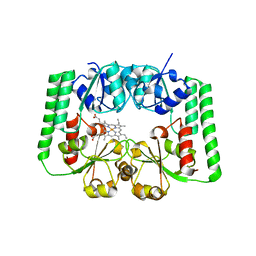 | |
3UBU
 
 | | Crystal structure of agkisacucetin, a GpIb-binding snaclec (snake C-type lectin) that inhibits platelet | | Descriptor: | Agglucetin subunit alpha-1, Agglucetin subunit beta-2, GLYCEROL, ... | | Authors: | Gao, Y, Ge, H, Chen, H, Li, H, Liu, Y, Niu, L, Teng, M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of agkisacucetin, a Gpib-binding snake C-type lectin that inhibits platelet adhesion and aggregation.
Proteins, 2012
|
|
1D0C
 
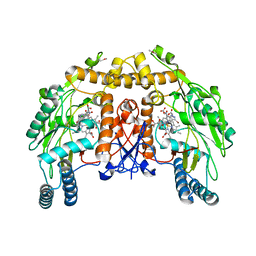 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 3-BROMO-7-NITROINDAZOLE (H4B FREE) | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, ACETATE ION, BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-09 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
6WPG
 
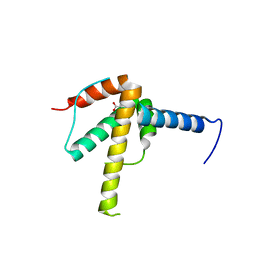 | | Structural Basis of Salicylic Acid Perception by Arabidopsis NPR Proteins | | Descriptor: | 2-HYDROXYBENZOIC ACID, Regulatory protein NPR4 | | Authors: | Wang, W, Withers, J, Li, H, Zwack, P.J, Rusnac, D.V, Shi, H, Liu, L, Yan, S, Hinds, T.R, Guttman, M, Dong, X, Zheng, N. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Structural basis of salicylic acid perception by Arabidopsis NPR proteins.
Nature, 586, 2020
|
|
6LLC
 
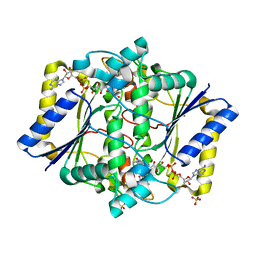 | | Discovery of A Dual Inhibitor of NQO1 and GSTP1 for Treating Malignant Glioblastoma | | Descriptor: | 5-methyl-N-(5-nitro-1,3-thiazol-2-yl)-3-phenyl-1,2-oxazole-4-carboxamide, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1, ... | | Authors: | Ye, K, Li, H. | | Deposit date: | 2019-12-23 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Discovery of a dual inhibitor of NQO1 and GSTP1 for treating glioblastoma.
J Hematol Oncol, 13, 2020
|
|
4R56
 
 | | Crystal structure of Sulfolobus Cren7-dsDNA(GTGATCAC) complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Chen, Y.Y, Li, H.B, Huang, L. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4
Extremophiles, 19, 2015
|
|
2QMX
 
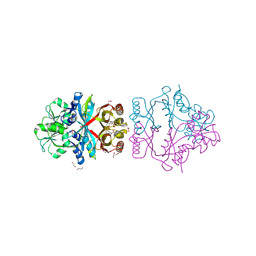 | | The crystal structure of L-Phe inhibited prephenate dehydratase from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHENYLALANINE, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
1ED4
 
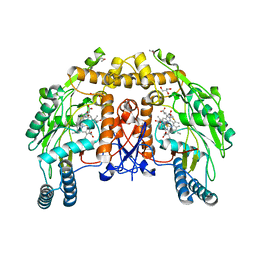 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH IPITU (H4B FREE) | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-01-26 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
4RGP
 
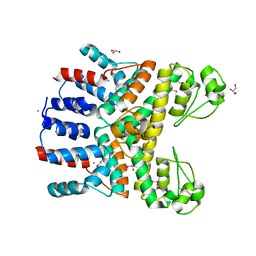 | | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans | | Descriptor: | CALCIUM ION, Csm6_III-A, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans
To be Published
|
|
1ED5
 
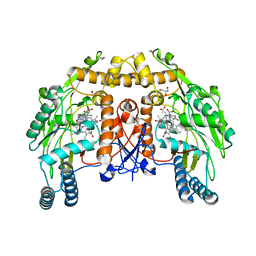 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH NNA(H4B FREE) | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-01-26 | | Release date: | 2001-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
4RNL
 
 | | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus | | Descriptor: | GLYCEROL, PHOSPHATE ION, possible galactose mutarotase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus
To be Published
|
|
2R79
 
 | | Crystal Structure of a Periplasmic Heme Binding Protein from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Periplasmic binding protein, ... | | Authors: | Ho, W.W, Li, H, Poulos, T.L. | | Deposit date: | 2007-09-07 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins.
J.Biol.Chem., 282, 2007
|
|
