7C8Z
 
 | |
5HAB
 
 | |
5IMU
 
 | | A fragment of conserved hypothetical protein Rv3899c (residues 184-410) from Mycobacterium tuberculosis | | Descriptor: | POTASSIUM ION, Tat (Twin-arginine translocation) pathway signal sequence containing protein | | Authors: | Li, D.F, Gao, Y.R, Liu, Y.Y, Bi, L.J. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rv3899c(184-410), a hypothetical protein from Mycobacterium tuberculosis
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
2DS2
 
 | | Crystal structure of mabinlin II | | Descriptor: | ACETIC ACID, Sweet protein mabinlin-2 chain A, Sweet protein mabinlin-2 chain B | | Authors: | Li, D.F, Zhu, D.Y, Wang, D.C. | | Deposit date: | 2006-06-19 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Mabinlin II: a novel structural type of sweet proteins and the main structural basis for its sweetness.
J.Struct.Biol., 162, 2008
|
|
8J65
 
 | |
2ZGQ
 
 | |
2ZGO
 
 | | Crystal structure of AAL mutant H59Q complex with lactose | | Descriptor: | Anti-tumor lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Li, D.F, Yang, N, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
2ZGM
 
 | | Crystal structure of recombinant Agrocybe aegerita lectin,rAAL, complex with lactose | | Descriptor: | Anti-tumor lectin, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Li, D.F, Yang, N, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
2ZGS
 
 | |
2ZGT
 
 | |
2ZGU
 
 | |
3AQT
 
 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | Descriptor: | Bacterial regulatory proteins, tetR family, RESORCINOL | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3AQS
 
 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3VSG
 
 | | Crystal structure of iron free 1,6-APD, 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSI
 
 | | Crystal structure of native 1,6-APD (2-Animophenol-1,6-dioxygenase) complex with 4-Nitrocatechol | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, 4-NITROCATECHOL, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSJ
 
 | | Crystal structure of 1,6-APD (2-ANIMOPHENOL-1,6-DIOXYGENASE) complexed with intermediate products | | Descriptor: | (2Z,4Z)-2-imino-6-oxohex-4-enoic acid, (3E)-3-iminooxepin-2(3H)-one, 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WE5
 
 | |
3VSH
 
 | | Crystal structure of native 1,6-APD (with Iron), 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, FE (II) ION | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6IYW
 
 | | Crystal sturucture of L,D-transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with Imipenem adduct | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, GLYCEROL, L,D-transpeptidase 2 | | Authors: | Li, D.F, Zhao, F, Wang, D.C. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1-beta-methyl group confers a lower affinity of l,d-transpeptidase LdtMt2 for ertapenem than for imipenem.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6LQB
 
 | | A third intermediate state of L,D-transpeptidase LdtMt2-ertapenem adduct | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, D.F, Qin, Y.L. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New State I-plus Observed for the L,D-transpeptidase LdtMt2-ertapenem Adduct
Prog.Biochem.Biophys., 47, 2020
|
|
5WS2
 
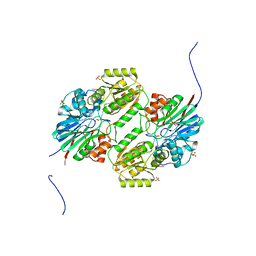 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, complex with RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*A)-3'), Ribonuclease J, SULFATE ION, ... | | Authors: | Li, D.F, Feng, N. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family
Mol. Microbiol., 106, 2017
|
|
6LLB
 
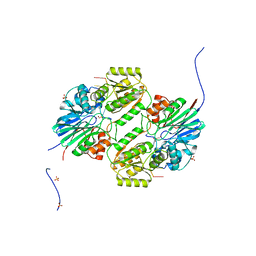 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, in complex with 6 nt RNA | | Descriptor: | MPY-RNase J, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, ... | | Authors: | Li, D.F, Hou, Y.J, Guo, L. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Rna Biol., 17, 2020
|
|
3TC1
 
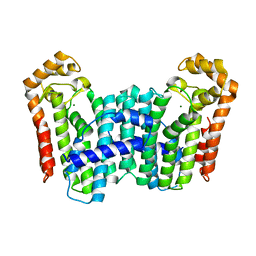 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori | | Descriptor: | MAGNESIUM ION, Octaprenyl Pyrophosphate Synthase | | Authors: | Zhang, J.Y, Zhang, X.L, Li, D.F, Zou, Q.M, Wang, D.C. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori
To be Published
|
|
8Y6T
 
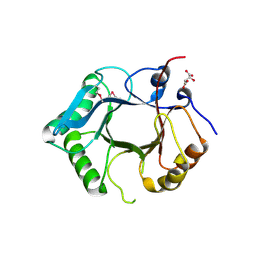 | |
5XUA
 
 | |
