6H1T
 
 | | Structure of the BM3 heme domain in complex with clotrimazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
2IUQ
 
 | |
5NX7
 
 | | Crystal structure of 1,8-cineole synthase from Streptomyces clavuligerus in complex with 2-fluoroneryl diphosphate and 2-fluorogeranyl diphosphate | | Descriptor: | (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
4ZAN
 
 | |
5NX5
 
 | | Crystal structure of Linalool/Nerolidol synthase from Streptomyces clavuligerus in complex with 2-fluorogeranyl diphosphate | | Descriptor: | (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
4ZAF
 
 | |
5LTM
 
 | |
4ZAG
 
 | |
5NX6
 
 | | Crystal structure of 1,8-cineole synthase from Streptomyces clavuligerus in complex with 2-fluoroneryl diphosphate | | Descriptor: | (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
6H6X
 
 | | Structure of an evolved dimeric form of the UbiD-class enzyme HmfF from Pelotomaculum thermopropionicum in complex with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3-polyprenyl-4-hydroxybenzoate decarboxylase and related decarboxylases, CALCIUM ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2018-07-30 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Enzymatic Carboxylation of 2-Furoic Acid Yields 2,5-Furandicarboxylic Acid (FDCA).
Acs Catalysis, 9, 2019
|
|
2IUV
 
 | |
2IUR
 
 | |
2HKM
 
 | |
2I0R
 
 | |
6R30
 
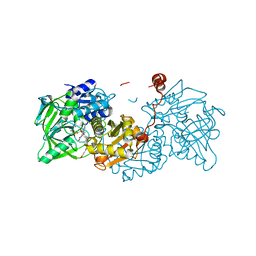 | |
6R3G
 
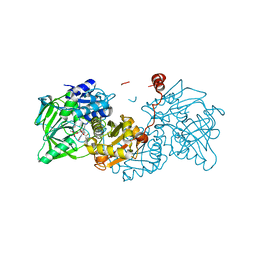 | |
6R3N
 
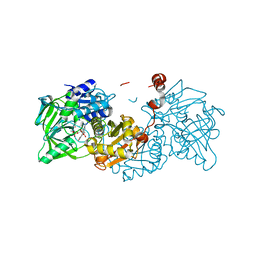 | |
6R33
 
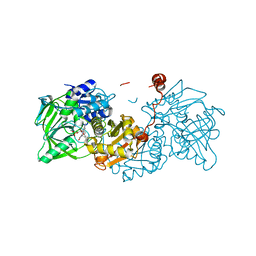 | |
6R3I
 
 | |
4ZAA
 
 | | Structure of A. niger Fdc1 in complex with 4-vinyl guaiacol | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, 2-Methoxy-4-vinylphenol, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
2HJB
 
 | |
4ZAD
 
 | | Structure of C. dubliensis Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAV
 
 | | UbiX in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4ZA8
 
 | | Crystal structure of A niger Fdc1 in complex with penta-fluorocinnamic acid | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, MANGANESE (II) ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAL
 
 | | Structure of UbiX E49Q mutant in complex with reduced FMN and dimethylallyl monophosphate | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dimethylallyl monophosphate, THIOCYANATE ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
