6WM6
 
 | | Periplasmic EDTA-binding protein EppA, tetragonal | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein, family 5, ... | | Authors: | Lewis, K.M, Sattler, S.A, Greene, C.L, Xun, L, Kang, C. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Structural Basis of the Binding of Various Aminopolycarboxylates by the Periplasmic EDTA-Binding Protein EppA from Chelativorans sp. BNC1.
Int J Mol Sci, 21, 2020
|
|
5KN1
 
 | | Recombinant bovine skeletal calsequestrin, high-Ca2+ form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lewis, K.M, Byrd, S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
6WM7
 
 | | Periplasmic EDTA-binding protein EppA, orthorhombic | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein, family 5, ... | | Authors: | Lewis, K.M, Greene, C.L, Sattler, S.A, Xun, L, Kang, C. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Structural Basis of the Binding of Various Aminopolycarboxylates by the Periplasmic EDTA-Binding Protein EppA from Chelativorans sp. BNC1.
Int J Mol Sci, 21, 2020
|
|
5KN0
 
 | | Native bovine skeletal calsequestrin, low-Ca2+ form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lewis, K.M, Byrd, S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
5KN2
 
 | | Native bovine skeletal calsequestrin, high-Ca2+ form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Calsequestrin | | Authors: | Lewis, K.M, Byrd, S.S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
5KN3
 
 | | Recombinant bovine skeletal calsequestrin, low-Ca2+ form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Lewis, K.M, Byrd, S, Kang, C. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Characterization of Post-Translational Modifications to Calsequestrins of Cardiac and Skeletal Muscle.
Int J Mol Sci, 17, 2016
|
|
5CRH
 
 | |
5CRG
 
 | |
5CRE
 
 | | Human skeletal calsequestrin, D210G mutant low-calcium complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
5CRD
 
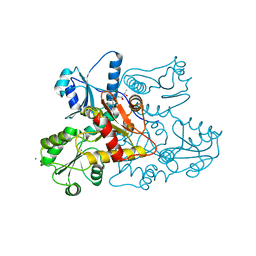 | | Wild-type human skeletal calsequestrin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
7UGW
 
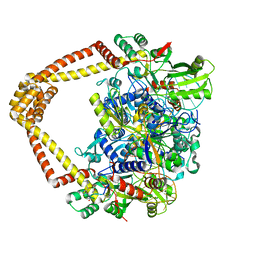 | | M. tuberculosis DNA gyrase cleavage core bound to DNA and evybactin | | Descriptor: | DNA (46-MER), DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Hauk, G, Imai, Y, Lewis, K, Berger, J.M. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evybactin is a DNA gyrase inhibitor that selectively kills Mycobacterium tuberculosis.
Nat.Chem.Biol., 18, 2022
|
|
6VBY
 
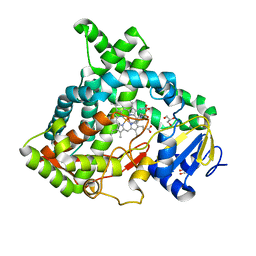 | | Cinnamate 4-hydroxylase (C4H1) from Sorghum bicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cinnamic acid 4-hydroxylase, GLYCEROL, ... | | Authors: | Zhang, B, Kang, C, Lewis, K.M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor.
Plant Physiol., 183, 2020
|
|
7T3H
 
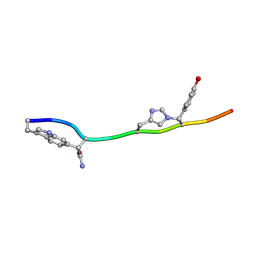 | | MicroED structure of Dynobactin | | Descriptor: | TRP-ASN-SER-ASN-VAL-HIS-SER-TYR-ARG-PHE | | Authors: | Yoo, B.-K, Kaiser, J.T, Rees, D.C, Miller, R.D, Iinishi, A, Lewis, K, Bowman, S. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
6PXS
 
 | | Crystal structure of iminodiacetate oxidase (IdaA) from Chelativorans sp. BNC1 | | Descriptor: | FAD dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Jun, S.Y, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structural and biochemical characterization of iminodiacetate oxidase from Chelativorans sp. BNC1.
Mol.Microbiol., 112, 2019
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRI
 
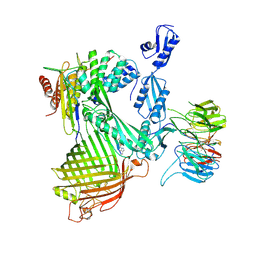 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
6P7K
 
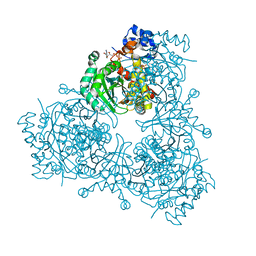 | | Structure of HMG-CoA reductase from Burkholderia cenocepacia | | Descriptor: | 3-hydroxy-3-methylglutaryl coenzyme A reductase, ADENOSINE-5'-DIPHOSPHATE, COENZYME A | | Authors: | Walker, A.M, Peacock, R.B, Hicks, C.W, Dewing, S.M, Lewis, K.M, Abboud, J, Stewart, S.W.A, Kang, C, Watson, J.M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Structural and Functional Characterization of Dynamic Oligomerization in Burkholderia cenocepacia HMG-CoA Reductase.
Biochemistry, 58, 2019
|
|
4LBP
 
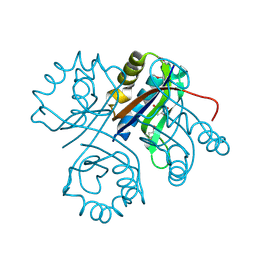 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Complex with 2,5-dihydroxybenzoquinone | | Descriptor: | 2,5-dihydroxycyclohexa-2,5-diene-1,4-dione, 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
4LBI
 
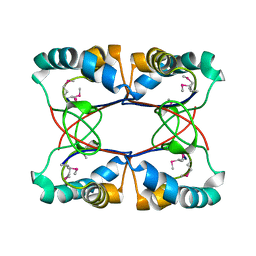 | |
4LBH
 
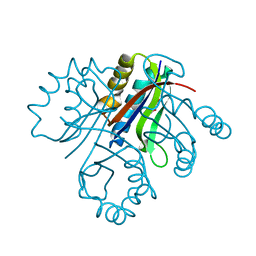 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Apo-form | | Descriptor: | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG), DI(HYDROXYETHYL)ETHER | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
4RPO
 
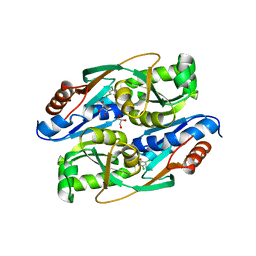 | | PcpR inducer binding domain (Complex with 2,4,6-trichlorophenol) | | Descriptor: | 2,4,6-trichlorophenol, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPN
 
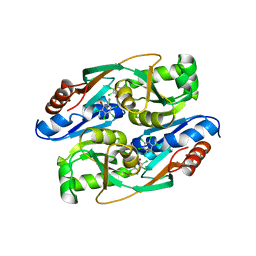 | | PcpR inducer binding domain complex with pentachlorophenol | | Descriptor: | PCP degradation transcriptional activation protein, PENTACHLOROPHENOL | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
3V1W
 
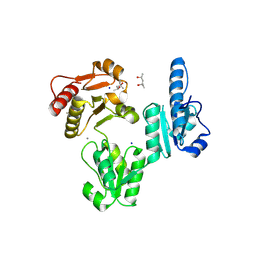 | | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Subramanian, A.K, Nissen, M.N, Lewis, K.M, Sanchez, E.J, Muralidharan, A.K, Kang, C. | | Deposit date: | 2011-12-10 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Molecular Basis for Multiple Ligand Binding of Calsequestrin and Potential Inhibition by Caffeine and Gallocatecin
To be Published
|
|
4RNS
 
 | | PcpR inducer binding domain (apo-form) | | Descriptor: | PCP degradation transcriptional activation protein | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
