1TJA
 
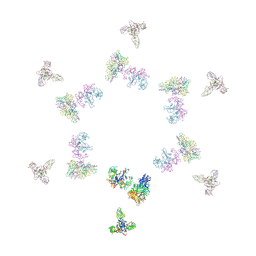 | | Fitting of gp8, gp9, and gp11 into the cryo-EM reconstruction of the bacteriophage T4 contracted tail | | Descriptor: | Baseplate structural protein Gp11, Baseplate structural protein Gp8, Baseplate structural protein Gp9 | | Authors: | Leiman, P.G, Chipman, P.R, Kostyuchenko, V.A, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2004-06-03 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Three-dimensional rearrangement of proteins in the tail of bacteriophage t4 on infection of its host
Cell(Cambridge,Mass.), 118, 2004
|
|
8VOL
 
 | |
8VOM
 
 | |
8VON
 
 | |
7UM0
 
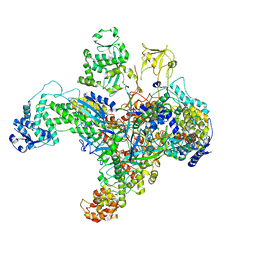 | | Structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with two DNA oligonucleotides containing the AR9 P077 promoter as determined by cryo-EM | | Descriptor: | DNA (5'-D(P*GP*UP*U)-3'), DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, ... | | Authors: | Leiman, P.G, Fraser, A, Sokolova, M.L. | | Deposit date: | 2022-04-05 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
7UM1
 
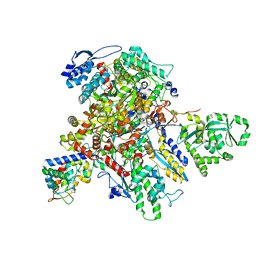 | | Structure of bacteriophage AR9 non-virion RNAP polymerase holoenzyme determined by cryo-EM | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Fraser, A, Sokolova, M.L. | | Deposit date: | 2022-04-05 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
5IW9
 
 | |
7S00
 
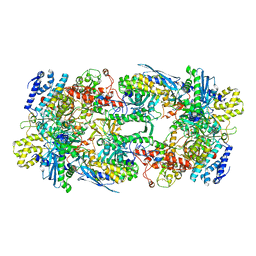 | | X-ray structure of the phage AR9 non-virion RNA polymerase core | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Sokolova, M.L, Fraser, A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
7S01
 
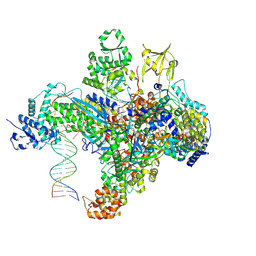 | | X-ray structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with a forked oligonucleotide containing the P077 promoter | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | Authors: | Leiman, P.G, Sokolova, M.L, Gordeeva, J, Fraser, A, Severinov, K.V. | | Deposit date: | 2021-08-28 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
1EL6
 
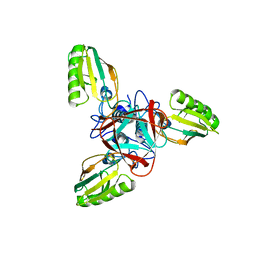 | | STRUCTURE OF BACTERIOPHAGE T4 GENE PRODUCT 11, THE INTERFACE BETWEEN THE BASEPLATE AND SHORT TAIL FIBERS | | Descriptor: | BASEPLATE STRUCTURAL PROTEIN GP11 | | Authors: | Leiman, P.G, Kostyuchenko, V.A, Schneider, M.M, Kurochkina, L.P, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2000-03-13 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of bacteriophage T4 gene product 11, the interface between the baseplate and short tail fibers.
J.Mol.Biol., 301, 2000
|
|
2FKK
 
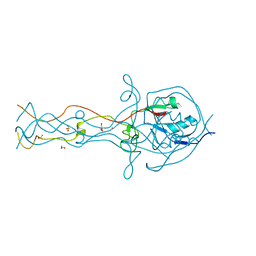 | | Crystal structure of the C-terminal domain of the bacteriophage T4 gene product 10 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Leiman, P.G, Shneider, M.M, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2006-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Evolution of bacteriophage tails: structure of t4 gene product 10
J.Mol.Biol., 358, 2006
|
|
2FL9
 
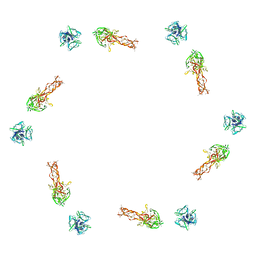 | |
2FL8
 
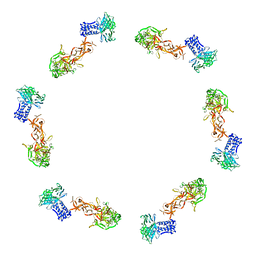 | |
6ORJ
 
 | |
6E0V
 
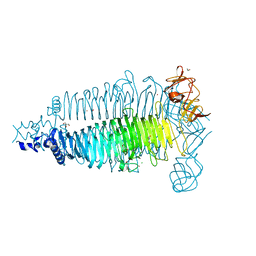 | | Apo crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriophage Phi92 gp150, CHLORIDE ION, ... | | Authors: | Leiman, P.G, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Plattner, M, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
6VR4
 
 | |
1N7Z
 
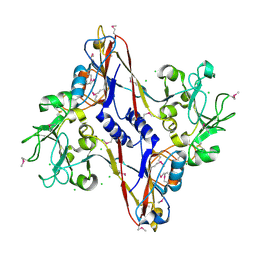 | | Structure and location of gene product 8 in the bacteriophage T4 baseplate | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N8B
 
 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | BROMIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
1N80
 
 | | Bacteriophage T4 baseplate structural protein gp8 | | Descriptor: | CHLORIDE ION, baseplate structural protein gp8 | | Authors: | Leiman, P.G, Shneider, M.M, Kostyuchenko, V.A, Chipman, P.R, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2002-11-18 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and location of gene product 8 in the bacteriophage T4 baseplate
J.Mol.Biol., 328, 2003
|
|
5LYE
 
 | |
5CES
 
 | |
3WIT
 
 | |
3PQI
 
 | |
4Y9V
 
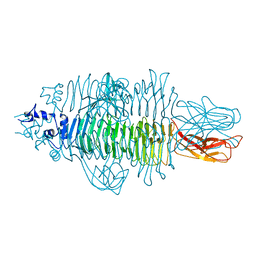 | | Gp54 tailspike of Acinetobacter baumannii bacteriophage AP22 in complex with A. baumannii capsular saccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2,4-dideoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-fucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-mannopyranuronic acid, CHLORIDE ION, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2015-02-17 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of Acinetobacter baumannii bacteriophage AP22 polysaccharide degrading lyase in complex with A. baumannii capsular saccharide at 0.9 A resolution
TO BE PUBLISHED
|
|
7CN7
 
 | |
