4JGS
 
 | |
7R22
 
 | |
1P26
 
 | | Crystal structure of zinc(II)-d(GGCGCC)2 | | Descriptor: | 5'-D(*GP*GP*CP*GP*CP*C)-3', ZINC ION | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
7S94
 
 | |
2AXY
 
 | | Crystal Structure of KH1 domain of human Poly(C)-binding protein-2 with C-rich strand of human telomeric DNA | | Descriptor: | C-rich strand of human telomeric dna, Poly(rC)-binding protein 2 | | Authors: | Du, Z, Lee, J.K, Tjhen, R.J, Li, S, Stroud, R.M, James, T.L. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the First KH Domain of Human Poly(C)-binding Protein-2 in Complex with a C-rich Strand of Human Telomeric DNA at 1.7 A
J.Biol.Chem., 280, 2005
|
|
3BVK
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
3BVL
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
1P24
 
 | | Crystal structure of cobalt(II)-d(GGCGCC)2 | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*GP*CP*GP*CP*C)-3') | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
3BPM
 
 | | Crystal Structure of Falcipain-3 with Its inhibitor, Leupeptin | | Descriptor: | Cysteine protease falcipain-3, Leupeptin, SULFATE ION | | Authors: | kerr, I.D, Lee, J.H, Brinen, L.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: implications for substrate specificity.
J.Med.Chem., 52, 2009
|
|
3BVI
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
1Q0K
 
 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the thiosulfate-reduced state | | Descriptor: | NICKEL (II) ION, SULFATE ION, Superoxide dismutase [Ni], ... | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3BVF
 
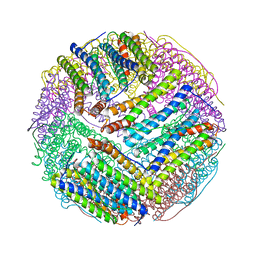 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
1N7E
 
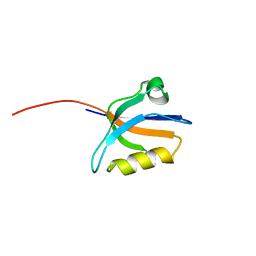 | | Crystal structure of the sixth PDZ domain of GRIP1 | | Descriptor: | AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
3BPF
 
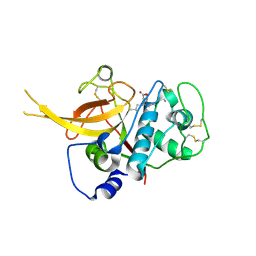 | | Crystal Structure of Falcipain-2 with Its inhibitor, E64 | | Descriptor: | Cysteine protease falcipain-2, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | kerr, I.D, Lee, J.H, Brinen, L.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: implications for substrate specificity.
J.Med.Chem., 52, 2009
|
|
6JZL
 
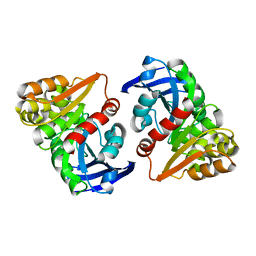 | |
4ERZ
 
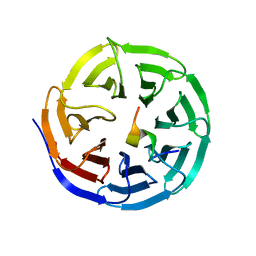 | | X-ray structure of WDR5-MLL4 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
2AZ0
 
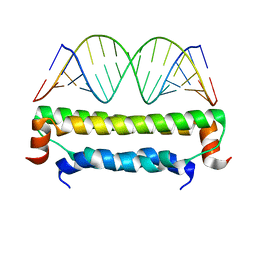 | | Flock House virus B2-dsRNA Complex (P212121) | | Descriptor: | 5'-R(*GP*CP*AP*(5BU)P*GP*GP*AP*CP*GP*CP*GP*(5BU)P*CP*CP*AP*(5BU)P*GP*C)-3', B2 protein | | Authors: | Chao, J.A, Lee, J.H, Chapados, B.R, Debler, E.W, Schneemann, A, Williamson, J.R. | | Deposit date: | 2005-09-09 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual modes of RNA-silencing suppression by Flock House virus protein B2.
Nat.Struct.Mol.Biol., 12, 2005
|
|
5WUX
 
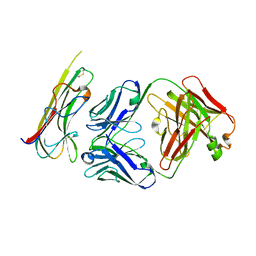 | | TNFalpha-certolizumab Fab | | Descriptor: | Tumor necrosis factor alpha, heavy, light | | Authors: | Heo, Y.S, Lee, J.U. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
1Q0G
 
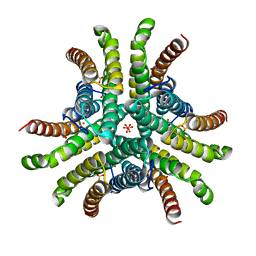 | | Crystal structure of Ni-containing superoxide dismutase with Ni-ligation corresponding to the state after full x-ray-induced reduction | | Descriptor: | NICKEL (II) ION, SULFATE ION, Superoxide dismutase [Ni] | | Authors: | Wuerges, J, Lee, J.-W, Yim, Y.-I, Yim, H.-S, Kang, S.-O, Djinovic Carugo, K. | | Deposit date: | 2003-07-16 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of nickel-containing superoxide dismutase reveals another type of active site
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3BVE
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
1I3C
 
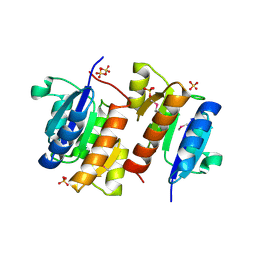 | | RESPONSE REGULATOR FOR CYANOBACTERIAL PHYTOCHROME, RCP1 | | Descriptor: | RESPONSE REGULATOR RCP1, SULFATE ION | | Authors: | Im, Y.J, Rho, S.-H, Park, C.-M, Yang, S.-S, Kang, J.-G, Lee, J.Y, Song, P.-S, Eom, S.H. | | Deposit date: | 2001-02-14 | | Release date: | 2002-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a cyanobacterial phytochrome response regulator.
Protein Sci., 11, 2002
|
|
4LVH
 
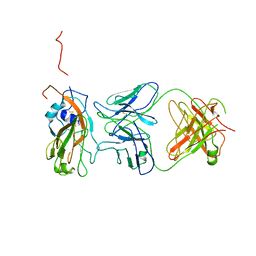 | | Insight into highly conserved H1 subtype-specific epitopes in influenza virus hemagglutinin | | Descriptor: | CALCIUM ION, Hemagglutinin, MONOCLONAL ANTIBODY H-CHAIN, ... | | Authors: | Kim, K.H, Cho, K.J, Kim, S, Seok, J.H, Lee, J.-H. | | Deposit date: | 2013-07-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight into highly conserved h1 subtype-specific epitopes in influenza virus hemagglutinin
Plos One, 9, 2014
|
|
1I7F
 
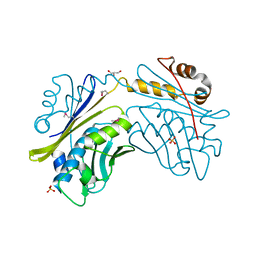 | | CRYSTAL STRUCTURE OF THE HSP33 DOMAIN WITH CONSTITUTIVE CHAPERONE ACTIVITY | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN 33, SULFATE ION | | Authors: | Kim, S.-J, Jeong, D.-G, Chi, S.-W, Lee, J.-S, Ryu, S.-E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-05-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity
Nat.Struct.Biol., 8, 2001
|
|
1P25
 
 | | Crystal structure of nickel(II)-d(GGCGCC)2 | | Descriptor: | 5'-D(*GP*GP*CP*GP*CP*C)-3', NICKEL (II) ION | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
2F0Y
 
 | |
